Cytotoxic effector functions of T cells are not required for protective immunity against fatal Rickettsia typhi infection in a murine model of infection: Role of TH1 and TH17 cytokines in protection and pathology
- PMID: 28222146
- PMCID: PMC5336310
- DOI: 10.1371/journal.pntd.0005404
Cytotoxic effector functions of T cells are not required for protective immunity against fatal Rickettsia typhi infection in a murine model of infection: Role of TH1 and TH17 cytokines in protection and pathology
Abstract
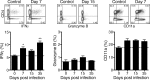
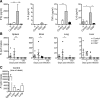
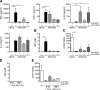
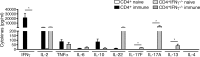
Endemic typhus caused by Rickettsia (R.) typhi is an emerging febrile disease that can be fatal due to multiple organ pathology. Here we analyzed the requirements for protection against R. typhi by T cells in the CB17 SCID model of infection. BALB/c wild-type mice generate CD4+ TH1 and cytotoxic CD8+ T cells both of which are sporadically reactivated in persistent infection. Either adoptively transferred CD8+ or CD4+ T cells protected R. typhi-infected CB17 SCID mice from death and provided long-term control. CD8+ T cells lacking either IFNγ or Perforin were still protective, demonstrating that the cytotoxic function of CD8+ T cells is not essential for protection. Immune wild-type CD4+ T cells produced high amounts of IFNγ, induced the release of nitric oxide in R. typhi-infected macrophages and inhibited bacterial growth in vitro via IFNγ and TNFα. However, adoptive transfer of CD4+IFNγ-/- T cells still protected 30-90% of R. typhi-infected CB17 SCID mice. These cells acquired a TH17 phenotype, producing high amounts of IL-17A and IL-22 in addition to TNFα, and inhibited bacterial growth in vitro. Surprisingly, the neutralization of either TNFα or IL-17A in CD4+IFNγ-/- T cell recipient mice did not alter bacterial elimination by these cells in vivo, led to faster recovery and enhanced survival compared to isotype-treated animals. Thus, collectively these data show that although CD4+ TH1 cells are clearly efficient in protection against R. typhi, CD4+ TH17 cells are similarly protective if the harmful effects of combined production of TNFα and IL-17A can be inhibited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Similar articles
-
CD4+ T Cells Are as Protective as CD8+ T Cells against Rickettsia typhi Infection by Activating Macrophage Bactericidal Activity.PLoS Negl Trop Dis. 2016 Nov 22;10(11):e0005089. doi: 10.1371/journal.pntd.0005089. eCollection 2016 Nov. PLoS Negl Trop Dis. 2016. PMID: 27875529 Free PMC article.
-
Liver Necrosis and Lethal Systemic Inflammation in a Murine Model of Rickettsia typhi Infection: Role of Neutrophils, Macrophages and NK Cells.PLoS Negl Trop Dis. 2016 Aug 22;10(8):e0004935. doi: 10.1371/journal.pntd.0004935. eCollection 2016 Aug. PLoS Negl Trop Dis. 2016. PMID: 27548618 Free PMC article.
-
GFPuv-Expressing Recombinant Rickettsia typhi: a Useful Tool for the Study of Pathogenesis and CD8+ T Cell Immunology in R. typhi Infection.Infect Immun. 2017 May 23;85(6):e00156-17. doi: 10.1128/IAI.00156-17. Print 2017 Jun. Infect Immun. 2017. PMID: 28289147 Free PMC article.
-
Dysregulation in lung immunity - the protective and pathologic Th17 response in infection.Eur J Immunol. 2013 Dec;43(12):3116-24. doi: 10.1002/eji.201343713. Epub 2013 Oct 16. Eur J Immunol. 2013. PMID: 24130019 Free PMC article. Review.
-
The Regulation of CD4(+) T Cell Responses during Protozoan Infections.Front Immunol. 2014 Oct 13;5:498. doi: 10.3389/fimmu.2014.00498. eCollection 2014. Front Immunol. 2014. PMID: 25352846 Free PMC article. Review.
Cited by
-
In silico construction of a multi-epitope vaccine (RGME-VAC/ATS-1) against the Rickettsia genus using immunoinformatics.Mem Inst Oswaldo Cruz. 2025 Mar 21;120:e240201. doi: 10.1590/0074-02760240201. eCollection 2025. Mem Inst Oswaldo Cruz. 2025. PMID: 40136144 Free PMC article.
-
The neglected challenge: Vaccination against rickettsiae.PLoS Negl Trop Dis. 2020 Oct 22;14(10):e0008704. doi: 10.1371/journal.pntd.0008704. eCollection 2020 Oct. PLoS Negl Trop Dis. 2020. PMID: 33091016 Free PMC article. Review.
-
Rickettsia Vaccine Candidate pVAX1-OmpB24 Stimulates TCD4+INF-γ+ and TCD8+INF-γ+ Lymphocytes in Autologous Co-Culture of Human Cells.Vaccines (Basel). 2023 Jan 13;11(1):173. doi: 10.3390/vaccines11010173. Vaccines (Basel). 2023. PMID: 36680017 Free PMC article.
-
Inflammatory cytokine profile and T cell responses in African tick bite fever patients.Med Microbiol Immunol. 2022 Jun;211(2-3):143-152. doi: 10.1007/s00430-022-00738-5. Epub 2022 May 11. Med Microbiol Immunol. 2022. PMID: 35543881 Free PMC article.
-
Vaccine Design and Vaccination Strategies against Rickettsiae.Vaccines (Basel). 2021 Aug 12;9(8):896. doi: 10.3390/vaccines9080896. Vaccines (Basel). 2021. PMID: 34452021 Free PMC article. Review.
References
-
- Rathi N, Rathi A. Rickettsial infections: Indian perspective. Indian Pediatr. 2010;47(2):157–64. Epub 2010/03/17. - PubMed
-
- Kuloglu F. Rickettsial infections. Disease and Molecular Medicine. 2013;1(2):39–45.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

