SlMAPK3 enhances tolerance to tomato yellow leaf curl virus (TYLCV) by regulating salicylic acid and jasmonic acid signaling in tomato (Solanum lycopersicum)
- PMID: 28222174
- PMCID: PMC5319765
- DOI: 10.1371/journal.pone.0172466
SlMAPK3 enhances tolerance to tomato yellow leaf curl virus (TYLCV) by regulating salicylic acid and jasmonic acid signaling in tomato (Solanum lycopersicum)
Abstract
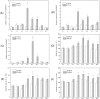
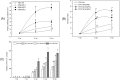
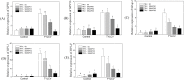
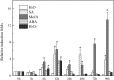
Several recent studies have reported on the role of mitogen-activated protein kinase (MAPK3) in plant immune responses. However, little is known about how MAPK3 functions in tomato (Solanum lycopersicum L.) infected with tomato yellow leaf curl virus (TYLCV). There is also uncertainty about the connection between plant MAPK3 and the salicylic acid (SA) and jasmonic acid (JA) defense-signaling pathways. The results of this study indicated that SlMAPK3 participates in the antiviral response against TYLCV. Tomato seedlings were inoculated with TYLCV to investigate the possible roles of SlMAPK1, SlMAPK2, and SlMAPK3 against this virus. Inoculation with TYLCV strongly induced the expression and the activity of all three genes. Silencing of SlMAPK1, SlMAPK2, and SlMAPK3 reduced tolerance to TYLCV, increased leaf H2O2 concentrations, and attenuated expression of defense-related genes after TYLCV infection, especially in SlMAPK3-silenced plants. Exogenous SA and methyl jasmonic acid (MeJA) both significantly induced SlMAPK3 expression in tomato leaves. Over-expression of SlMAPK3 increased the transcript levels of SA/JA-mediated defense-related genes (PR1, PR1b/SlLapA, SlPI-I, and SlPI-II) and enhanced tolerance to TYLCV. After TYLCV inoculation, the leaves of SlMAPK3 over-expressed plants compared with wild type plants showed less H2O2 accumulation and greater superoxide dismutase (SOD), peroxidase (POD), catalase (CAT), and ascorbate peroxidase (APX) activity. Overall, the results suggested that SlMAPK3 participates in the antiviral response of tomato to TYLCV, and that this process may be through either the SA or JA defense-signaling pathways.
Conflict of interest statement
Figures

Similar articles
-
A novel plant protein-disulfide isomerase participates in resistance response against the TYLCV in tomato.Planta. 2020 Jul 17;252(2):25. doi: 10.1007/s00425-020-03430-1. Planta. 2020. PMID: 32681182
-
Infection of tomato plants by tomato yellow leaf curl virus (TYLCV) potentiates the ethylene and salicylic acid pathways to fend off root-knot nematode (Meloidogyne incognita) parasitism.Plant Physiol Biochem. 2024 Dec;217:109271. doi: 10.1016/j.plaphy.2024.109271. Epub 2024 Nov 4. Plant Physiol Biochem. 2024. PMID: 39504658
-
Eugenol confers resistance to Tomato yellow leaf curl virus (TYLCV) by regulating the expression of SlPer1 in tomato plants.N Biotechnol. 2016 May 25;33(3):345-54. doi: 10.1016/j.nbt.2016.01.001. Epub 2016 Jan 14. N Biotechnol. 2016. PMID: 26776605
-
Knockout of SlMAPK3 Reduced Disease Resistance to Botrytis cinerea in Tomato Plants.J Agric Food Chem. 2018 Aug 29;66(34):8949-8956. doi: 10.1021/acs.jafc.8b02191. Epub 2018 Aug 20. J Agric Food Chem. 2018. PMID: 30092129
-
Exploiting Virus Infection to Protect Plants from Abiotic Stresses: Tomato Protection by a Begomovirus.Plants (Basel). 2022 Nov 1;11(21):2944. doi: 10.3390/plants11212944. Plants (Basel). 2022. PMID: 36365396 Free PMC article. Review.
Cited by
-
Evolutionarily conserved 12-oxophytodienoate reductase trans-lncRNA pair affects disease resistance in tea (Camellia sinensis) via the jasmonic acid signaling pathway.Hortic Res. 2024 May 6;11(7):uhae129. doi: 10.1093/hr/uhae129. eCollection 2024 Jul. Hortic Res. 2024. PMID: 38966865 Free PMC article.
-
Plant Defense and Viral Counter-Defense during Plant-Geminivirus Interactions.Viruses. 2023 Feb 12;15(2):510. doi: 10.3390/v15020510. Viruses. 2023. PMID: 36851725 Free PMC article. Review.
-
Interaction between SlMAPK3 and SlASR4 regulates drought resistance in tomato (Solanum lycopersicum L.).Mol Breed. 2023 Oct 3;43(10):73. doi: 10.1007/s11032-023-01418-9. eCollection 2023 Oct. Mol Breed. 2023. PMID: 37795156 Free PMC article.
-
Plant Mitogen-Activated Protein Kinase Cascades in Environmental Stresses.Int J Mol Sci. 2021 Feb 3;22(4):1543. doi: 10.3390/ijms22041543. Int J Mol Sci. 2021. PMID: 33546499 Free PMC article. Review.
-
Physiological and transcriptomic analyses revealed gene networks involved in heightened resistance against tomato yellow leaf curl virus infection in salicylic acid and jasmonic acid treated tomato plants.Front Microbiol. 2022 Sep 14;13:970139. doi: 10.3389/fmicb.2022.970139. eCollection 2022. Front Microbiol. 2022. PMID: 36187991 Free PMC article.
References
-
- Palukaitis P, Carr J. Plant resistance responses to viruses. Journal of Plant Pathology. 2008:153–71.
-
- Zhao PP, Shang J, Guo ZC, Xie HF, Xi DH, Sun X, et al. Temperature-related effects of treatments with jasmonic and salicylic acids on Arabidopsis infected with cucumber mosaic virus. Russian Journal of Plant Physiology. 2013; (No.5): 672–680
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

