Abnormal epigenetic changes during differentiation of human skeletal muscle stem cells from obese subjects
- PMID: 28222718
- PMCID: PMC5320752
- DOI: 10.1186/s12916-017-0792-x
Abnormal epigenetic changes during differentiation of human skeletal muscle stem cells from obese subjects
Abstract
Background: Human skeletal muscle stem cells are important for muscle regeneration. However, the combined genome-wide DNA methylation and expression changes taking place during adult myogenesis have not been described in detail and novel myogenic factors may be discovered. Additionally, obesity is associated with low relative muscle mass and diminished metabolism. Epigenetic alterations taking place during myogenesis might contribute to these defects.
Methods: We used Infinium HumanMethylation450 BeadChip Kit (Illumina) and HumanHT-12 Expression BeadChip (Illumina) to analyze genome-wide DNA methylation and transcription before versus after differentiation of primary human myoblasts from 14 non-obese and 14 obese individuals. Functional follow-up experiments were performed using siRNA mediated gene silencing in primary human myoblasts and a transgenic mouse model.
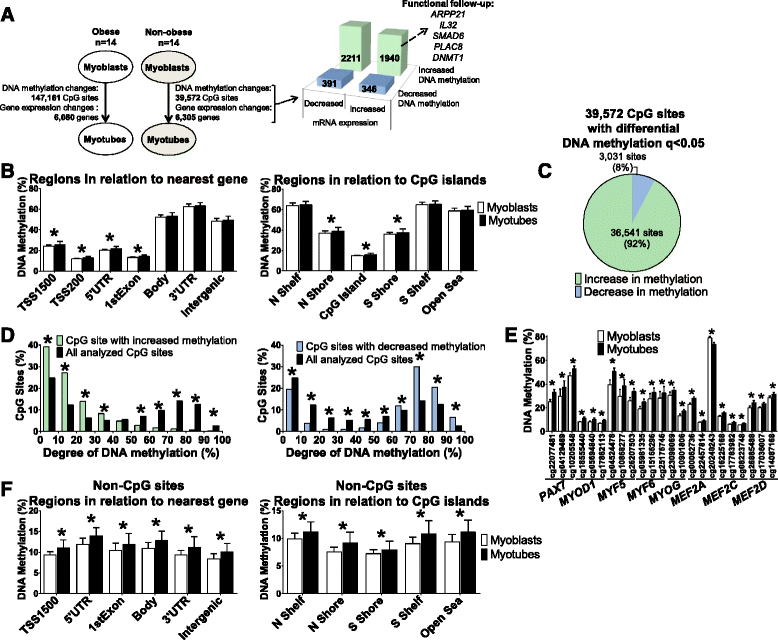
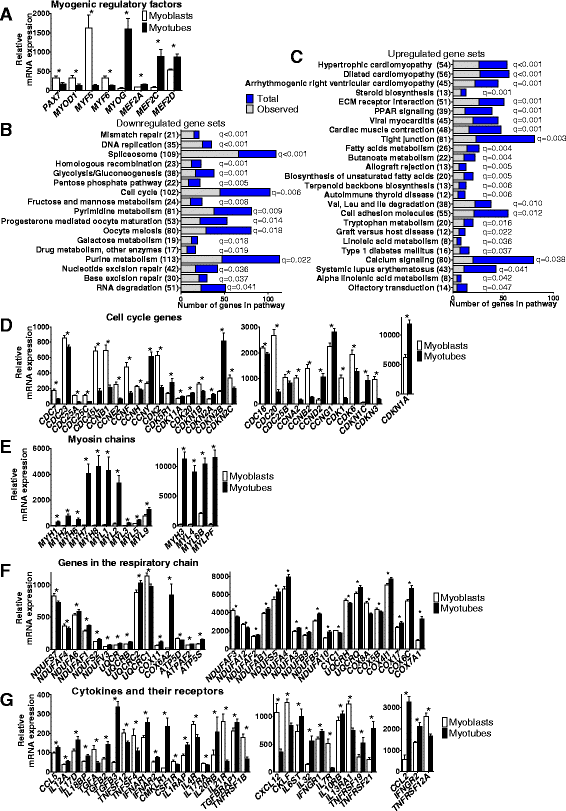
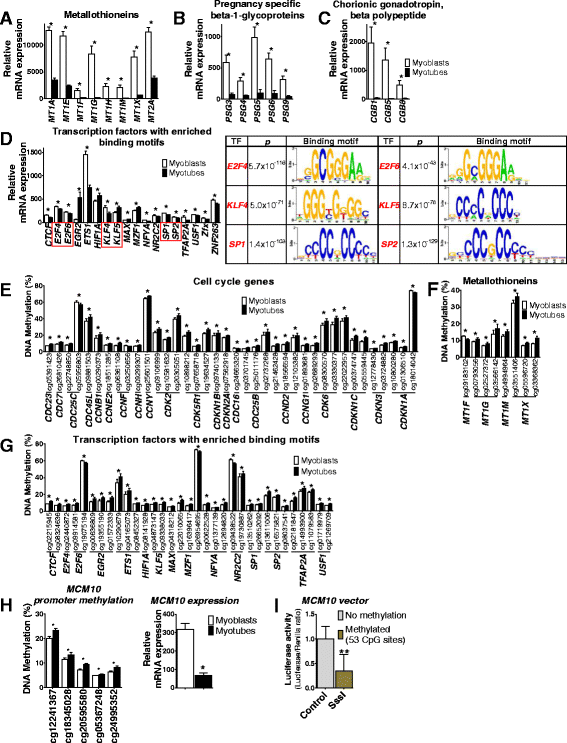
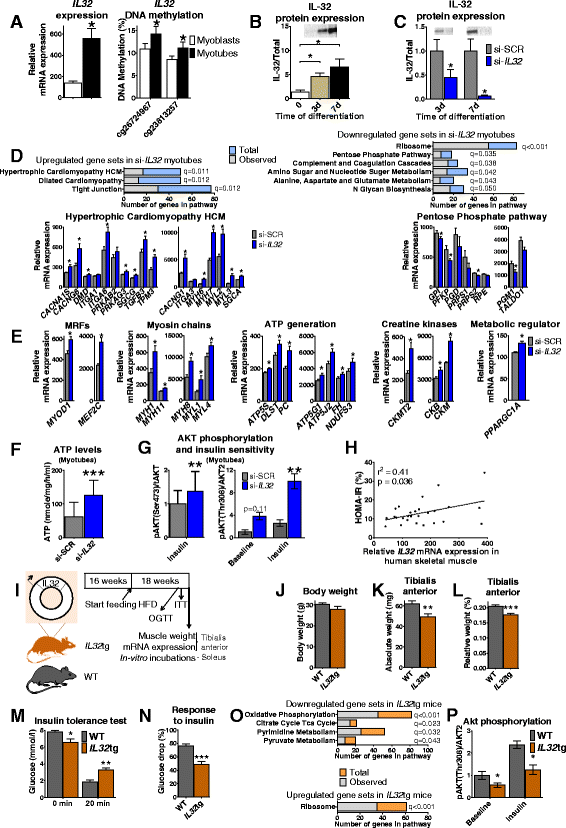
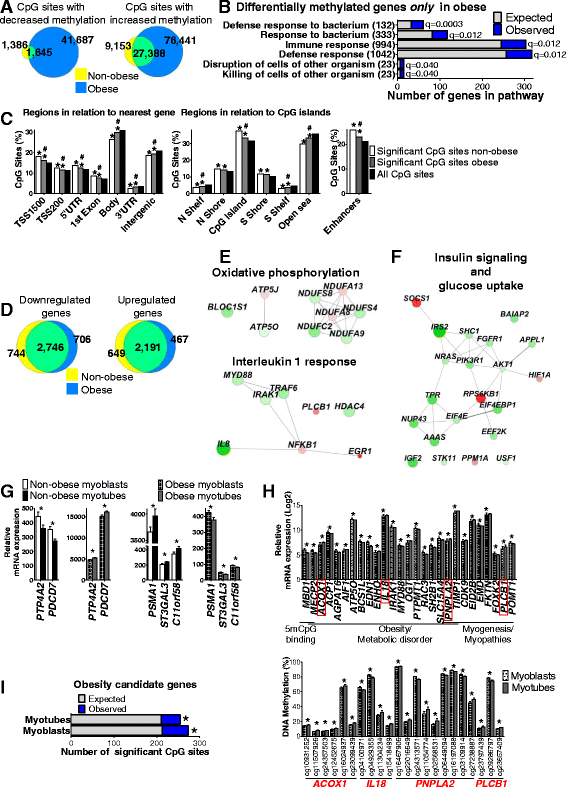
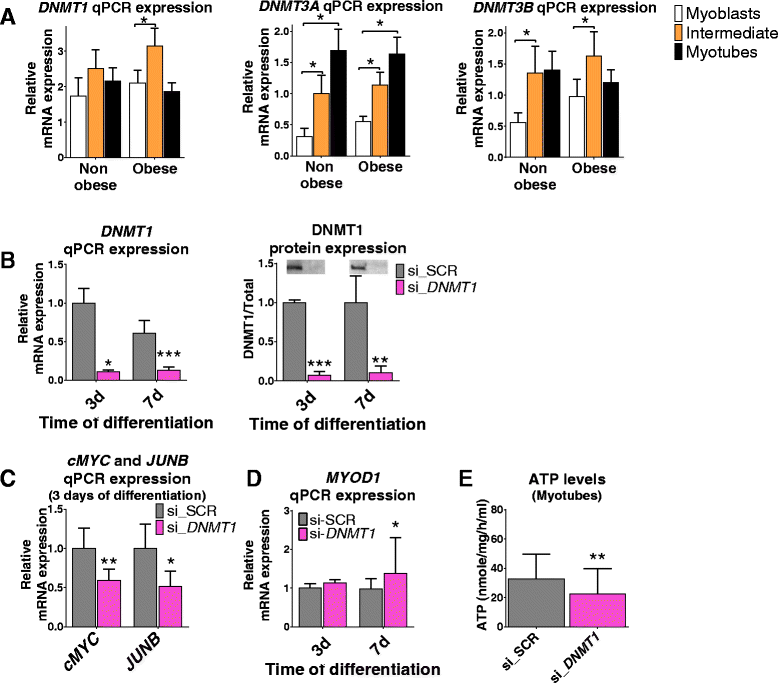
Results: We observed genome-wide changes in DNA methylation and expression patterns during differentiation of primary human muscle stem cells (myoblasts). We identified epigenetic and transcriptional changes of myogenic transcription factors (MYOD1, MYOG, MYF5, MYF6, PAX7, MEF2A, MEF2C, and MEF2D), cell cycle regulators, metabolic enzymes and genes previously not linked to myogenesis, including IL32, metallothioneins, and pregnancy-specific beta-1-glycoproteins. Functional studies demonstrated IL-32 as a novel target that regulates human myogenesis, insulin sensitivity and ATP levels in muscle cells. Furthermore, IL32 transgenic mice had reduced insulin response and muscle weight. Remarkably, approximately 3.7 times more methylation changes (147,161 versus 39,572) were observed during differentiation of myoblasts from obese versus non-obese subjects. In accordance, DNMT1 expression increased during myogenesis only in obese subjects. Interestingly, numerous genes implicated in metabolic diseases and epigenetic regulation showed differential methylation and expression during differentiation only in obese subjects.
Conclusions: Our study identifies IL-32 as a novel myogenic regulator, provides a comprehensive map of the dynamic epigenome during differentiation of human muscle stem cells and reveals abnormal epigenetic changes in obesity.
Keywords: ARPP21; CGB; DNA methylation; Epigenetics; IL-32; Insulin resistance; MT; Myogenesis; Obesity; PSG; TGF-β3.
Figures
Similar articles
-
Deconstruction of DNA methylation patterns during myogenesis reveals specific epigenetic events in the establishment of the skeletal muscle lineage.Stem Cells. 2015 Jun;33(6):2025-36. doi: 10.1002/stem.1998. Epub 2015 Apr 21. Stem Cells. 2015. PMID: 25801824
-
Dnmt1 regulates the myogenic lineage specification of muscle stem cells.Sci Rep. 2016 Oct 18;6:35355. doi: 10.1038/srep35355. Sci Rep. 2016. PMID: 27752090 Free PMC article.
-
DNA methylation analysis of human myoblasts during in vitro myogenic differentiation: de novo methylation of promoters of muscle-related genes and its involvement in transcriptional down-regulation.Hum Mol Genet. 2015 Jan 15;24(2):410-23. doi: 10.1093/hmg/ddu457. Epub 2014 Sep 4. Hum Mol Genet. 2015. PMID: 25190712 Free PMC article.
-
Concise Review: Epigenetic Regulation of Myogenesis in Health and Disease.Stem Cells Transl Med. 2016 Mar;5(3):282-90. doi: 10.5966/sctm.2015-0266. Epub 2016 Jan 21. Stem Cells Transl Med. 2016. PMID: 26798058 Free PMC article. Review.
-
Epigenetic Regulation of Adult Myogenesis.Curr Top Dev Biol. 2018;126:235-284. doi: 10.1016/bs.ctdb.2017.08.002. Epub 2017 Sep 28. Curr Top Dev Biol. 2018. PMID: 29305001 Review.
Cited by
-
Dnmt3b Deficiency in Myf5+-Brown Fat Precursor Cells Promotes Obesity in Female Mice.Biomolecules. 2021 Jul 23;11(8):1087. doi: 10.3390/biom11081087. Biomolecules. 2021. PMID: 34439754 Free PMC article.
-
Metabolic memory: mechanisms and diseases.Signal Transduct Target Ther. 2024 Feb 28;9(1):38. doi: 10.1038/s41392-024-01755-x. Signal Transduct Target Ther. 2024. PMID: 38413567 Free PMC article. Review.
-
A reference single-cell transcriptomic atlas of human skeletal muscle tissue reveals bifurcated muscle stem cell populations.Skelet Muscle. 2020 Jul 6;10(1):19. doi: 10.1186/s13395-020-00236-3. Skelet Muscle. 2020. PMID: 32624006 Free PMC article.
-
Characterization of Skeletal Muscle Endocrine Control in an In Vitro Model of Myogenesis.Calcif Tissue Int. 2020 Jul;107(1):18-30. doi: 10.1007/s00223-020-00678-3. Epub 2020 Feb 27. Calcif Tissue Int. 2020. PMID: 32107602 Free PMC article.
-
Immune modulation and prognostic significance of MCM10 in pan-cancer: a comprehensive analysis.Am J Transl Res. 2023 Nov 15;15(11):6451-6463. eCollection 2023. Am J Transl Res. 2023. PMID: 38074804 Free PMC article.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

