New H6 influenza virus reassortment strains isolated from Anser fabalis in Anhui Province, China
- PMID: 28222765
- PMCID: PMC5320792
- DOI: 10.1186/s12985-017-0680-1
New H6 influenza virus reassortment strains isolated from Anser fabalis in Anhui Province, China
Abstract
Background: H6 subtype avian influenza viruses are globally distributed and, in recent years, have been isolated with increasing frequency from both domestic and wild bird species as well as infected humans. Many reports have examined the viruses in the context of poultry or several wild bird species, but there is less information regarding their presence in migratory birds.
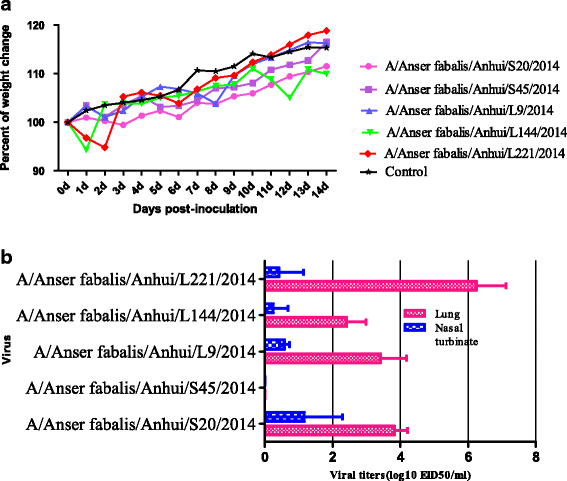
Methods: Hemagglutination and hemagglutination inhibition tests were used to measure HA activity for different HA subtypes. Whole viral genomes were sequenced and analysed using DNAstar and MEGA 6 to understand their genetic evolution. Pathogenicity was evaluated using a mouse infection model.
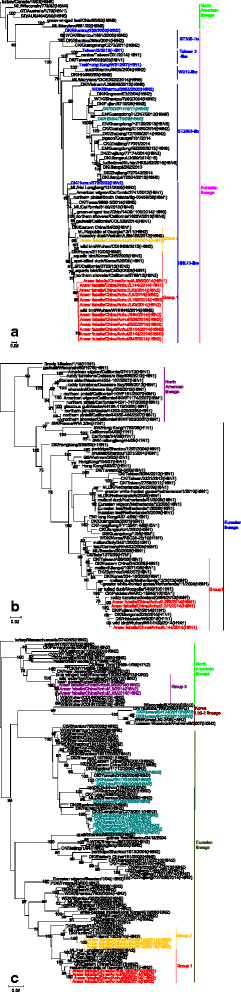
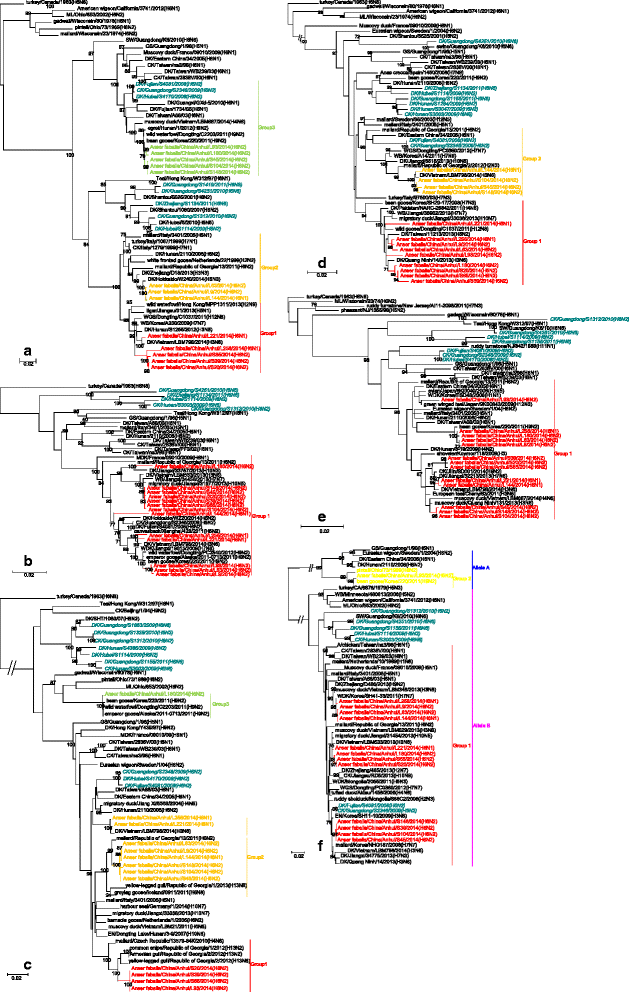
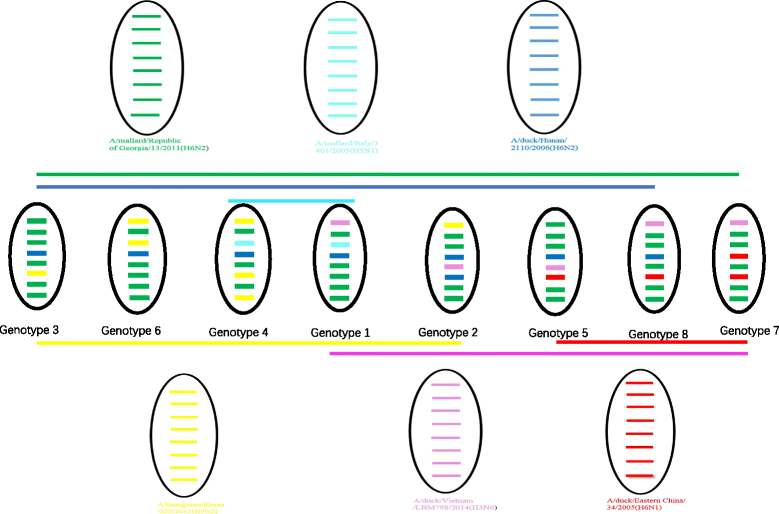
Results: We isolated 13 strains of H6 virus from faecal samples of migratory waterfowl in Anhui Province of China in 2014. Phylogenetic analysis showed gene reassortment between Eurasian and North American lineages. Five of the identified H6 strains had the ability to infect mice without adaptation.
Conclusion: Our findings suggest that regular surveillance of wild birds, especially migratory birds, is important for providing early warning and control of avian influenza outbreaks.
Keywords: Anser fabalis; Avian influenza virus; H6 subtype.
Figures
References
-
- Lewis NS, Verhagen JH, Javakhishvili Z, Russell CA, Lexmond P, Westgeest KB, et al. Influenza A virus evolution and spatio-temporal dynamics in Eurasian wild birds: a phylogenetic and phylogeographical study of whole-genome sequence data. J Gen Virol. 2015;96:2050–60. doi: 10.1099/vir.0.000155. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

