Antimicrobial resistance and genomic rep-PCR fingerprints of Pseudomonas aeruginosa strains from animals on the background of the global population structure
- PMID: 28222788
- PMCID: PMC5319083
- DOI: 10.1186/s12917-017-0977-8
Antimicrobial resistance and genomic rep-PCR fingerprints of Pseudomonas aeruginosa strains from animals on the background of the global population structure
Abstract
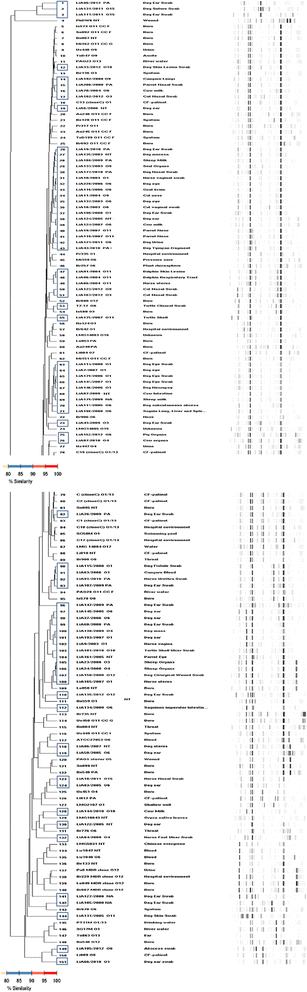
Background: Pseudomonas aeruginosa is an important human opportunistic pathogen responsible for fatal nosocomial infections worldwide, and has emerged as a relevant animal pathogen. Treatment options are dramatically decreasing, due to antimicrobial resistance and the microorganism's large versatile genome. Antimicrobial resistance profiles, serotype frequency and genomic profile of unrelated P. aeruginosa isolates of veterinary origin (n = 73), including domesticated, farm, zoo and wild animals mainly from Portugal were studied. The genomic profile, determined by DiversiLab system (Rep-PCR-based technique), was compared with the P. aeruginosa global population structure to evaluate their relatedness.
Results: Around 40% of the isolates expressed serotypes O6 (20.5%) and O1 (17.8%). A total of 46.6% of isolates was susceptible to all antimicrobials tested. Isolates obtained from most animals were non-multidrug resistant (86.3%), whereas 11% were multidrug resistant, MDR (non-susceptible to at least one agent in ≥ three antimicrobial categories), and 2.7% extensively drug resistant, XDR (non-susceptible to at least one agent in all but two or fewer antimicrobial categories). Resistance percentages were as follows: amikacin (0.0%), aztreonam (41.1%), cefepime (9.6%), ceftazidime (2.7%), ciprofloxacin (15.1%), colistin (0.0%), gentamicin (12.3%), imipenem (1.4%), meropenem (1.4%), piperacillin + tazobactam (12.3%), ticarcillin (16.4%), ticarcillin + clavulanic acid (17.8%), and tobramycin (1.4%). Animal isolates form a population with a non-clonal epidemic structure indistinguishable from the global P. aeruginosa population structure, where no specific 'animal clonal lineage' was detected.
Conclusions: Serotypes O6 and O1 were the most frequent. Serotype frequency and antimicrobial resistance patterns found in P. aeruginosa from animals were as expected for this species. This study confirms earlier results that P. aeruginosa has a non-clonal population structure, and shows that P. aeruginosa population from animals is homogeneously scattered and indistinguishable from the global population structure.
Keywords: Animal origin; Antimicrobials; Environment; MDR; Non-clonal; Population; Pseudomonas aeruginosa; Rep-PCR; Serotype; XDR.
Figures
Similar articles
-
Activity of ceftolozane/tazobactam against a collection of Pseudomonas aeruginosa isolates from bloodstream infections in Australia.Pathology. 2018 Dec;50(7):748-752. doi: 10.1016/j.pathol.2018.08.009. Epub 2018 Nov 2. Pathology. 2018. PMID: 30392710
-
Genetic fingerprinting and antimicrobial susceptibility profiles of Pseudomonas aeruginosa hospital isolates in Malaysia.J Microbiol Immunol Infect. 2009 Jun;42(3):197-209. J Microbiol Immunol Infect. 2009. PMID: 19812853
-
The identification, typing, and antimicrobial susceptibility of Pseudomonas aeruginosa isolated from mink with hemorrhagic pneumonia.Vet Microbiol. 2014 Jun 4;170(3-4):456-61. doi: 10.1016/j.vetmic.2014.02.025. Epub 2014 Feb 25. Vet Microbiol. 2014. PMID: 24629901
-
Pooled prevalence and trends of antimicrobial resistance in Pseudomonas aeruginosa clinical isolates over the past 10 years in Turkey: A meta-analysis.J Glob Antimicrob Resist. 2019 Sep;18:64-70. doi: 10.1016/j.jgar.2019.01.032. Epub 2019 Feb 10. J Glob Antimicrob Resist. 2019. PMID: 30753904
-
Age of Antibiotic Resistance in MDR/XDR Clinical Pathogen of Pseudomonas aeruginosa.Pharmaceuticals (Basel). 2023 Aug 30;16(9):1230. doi: 10.3390/ph16091230. Pharmaceuticals (Basel). 2023. PMID: 37765038 Free PMC article. Review.
Cited by
-
Advanced DNA fingerprint genotyping based on a model developed from real chip electrophoresis data.J Adv Res. 2019 Jan 25;18:9-18. doi: 10.1016/j.jare.2019.01.005. eCollection 2019 Jul. J Adv Res. 2019. PMID: 30788173 Free PMC article.
-
Molecular Detection and Phylogenetic Analysis of Pseudomonas aeruginosa Isolated from Some Infected and Healthy Ruminants in Basrah, Iraq.Arch Razi Inst. 2022 Apr 30;77(2):537-544. doi: 10.22092/ARI.2022.357802.2099. eCollection 2022 Apr. Arch Razi Inst. 2022. PMID: 36284961 Free PMC article.
-
Antibiotic Resistance and Virulence Profiles of Gram-Negative Bacteria Isolated from Loggerhead Sea Turtles (Caretta caretta) of the Island of Maio, Cape Verde.Antibiotics (Basel). 2021 Jun 24;10(7):771. doi: 10.3390/antibiotics10070771. Antibiotics (Basel). 2021. PMID: 34202799 Free PMC article.
-
Distribution and Characteristics of Bacteria Isolated from Cystic Fibrosis Patients with Pulmonary Exacerbation.Can J Infect Dis Med Microbiol. 2022 Dec 24;2022:5831139. doi: 10.1155/2022/5831139. eCollection 2022. Can J Infect Dis Med Microbiol. 2022. PMID: 36593975 Free PMC article.
-
Comparing antibiotic resistance and virulence profiles of Enterococcus faecium, Klebsiella pneumoniae, and Pseudomonas aeruginosa from environmental and clinical settings.Heliyon. 2024 Apr 26;10(9):e30215. doi: 10.1016/j.heliyon.2024.e30215. eCollection 2024 May 15. Heliyon. 2024. PMID: 38720709 Free PMC article.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

