Peptide recognition by heterochromatin protein 1 (HP1) chromoshadow domains revisited: Plasticity in the pseudosymmetric histone binding site of human HP1
- PMID: 28223359
- PMCID: PMC5392561
- DOI: 10.1074/jbc.M116.768374
Peptide recognition by heterochromatin protein 1 (HP1) chromoshadow domains revisited: Plasticity in the pseudosymmetric histone binding site of human HP1
Abstract
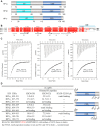
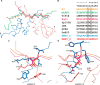
Heterochromatin protein 1 (HP1), a highly conserved non-histone chromosomal protein in eukaryotes, plays important roles in the regulation of gene transcription. Each of the three human homologs of HP1 includes a chromoshadow domain (CSD). The CSD interacts with various proteins bearing the PXVXL motif but also with a region of histone H3 that bears the similar PXXVXL motif. The latter interaction has not yet been resolved in atomic detail. Here we demonstrate that the CSDs of all three human HP1 homologs have comparable affinities to the PXXVXL motif of histone H3. The HP1 C-terminal extension enhances the affinity, as does the increasing length of the H3 peptide. The crystal structure of the human HP1γ CSD (CSDγ) in complex with an H3 peptide suggests that recognition of H3 by CSDγ to some extent resembles CSD-PXVXL interaction. Nevertheless, the prolyl residue of the PXXVXL motif appears to play a role distinct from that of Pro in the known HP1β CSD-PXVXL complexes. We consequently generalize the historical CSD-PXVXL interaction model and expand the search scope for additional CSD binding partners.
Keywords: chromatin regulation; chromatin structure; heterochromatin; histone; peptide interaction.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



References
-
- Will H., and Bautz E. K. F. (1980) Immunological identification of a chromocenter-associated protein in polytene chromosomes of Drosophila. Exp. Cell Res. 125, 401–410 - PubMed
-
- Eissenberg J. C., and Elgin S. C. (2000) The HP1 protein family: getting a grip on chromatin. Curr. Opin. Genet. Dev. 10, 204–210 - PubMed
-
- Maison C., and Almouzni G. (2004) HP1 and the dynamics of heterochromatin maintenance. Nat. Rev. Mol. Cell Biol. 5, 296–304 - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

