PEMapper and PECaller provide a simplified approach to whole-genome sequencing
- PMID: 28223510
- PMCID: PMC5347547
- DOI: 10.1073/pnas.1618065114
PEMapper and PECaller provide a simplified approach to whole-genome sequencing
Abstract
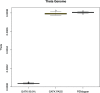
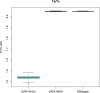
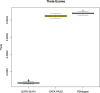
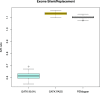
The analysis of human whole-genome sequencing data presents significant computational challenges. The sheer size of datasets places an enormous burden on computational, disk array, and network resources. Here, we present an integrated computational package, PEMapper/PECaller, that was designed specifically to minimize the burden on networks and disk arrays, create output files that are minimal in size, and run in a highly computationally efficient way, with the single goal of enabling whole-genome sequencing at scale. In addition to improved computational efficiency, we implement a statistical framework that allows for a base by base error model, allowing this package to perform as well or better than the widely used Genome Analysis Toolkit (GATK) in all key measures of performance on human whole-genome sequences.
Keywords: GATK; SNP calling; genome sequencing; sequence mapping; software.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Comment in
-
Reply to Plüss et al.: The strength of PEMapper/PECaller lies in unbiased calling using large sample sizes.Proc Natl Acad Sci U S A. 2017 Oct 3;114(40):E8323. doi: 10.1073/pnas.1714535114. Epub 2017 Sep 15. Proc Natl Acad Sci U S A. 2017. PMID: 28916730 Free PMC article. No abstract available.
-
Need for speed in accurate whole-genome data analysis: GENALICE MAP challenges BWA/GATK more than PEMapper/PECaller and Isaac.Proc Natl Acad Sci U S A. 2017 Oct 3;114(40):E8320-E8322. doi: 10.1073/pnas.1713830114. Epub 2017 Sep 15. Proc Natl Acad Sci U S A. 2017. PMID: 28916731 Free PMC article. No abstract available.
References
-
- Levy SE, Myers RM. Advancements in next-generation sequencing. Annu Rev Genomics Hum Genet. 2016;17:95–115. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

