Diagnostic exome sequencing in 266 Dutch patients with visual impairment
- PMID: 28224992
- PMCID: PMC5437915
- DOI: 10.1038/ejhg.2017.9
Diagnostic exome sequencing in 266 Dutch patients with visual impairment
Abstract
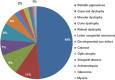
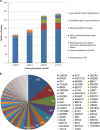
Inherited eye disorders have a large clinical and genetic heterogeneity, which makes genetic diagnosis cumbersome. An exome-sequencing approach was developed in which data analysis was divided into two steps: the vision gene panel and exome analysis. In the vision gene panel analysis, variants in genes known to cause inherited eye disorders were assessed for pathogenicity. If no causative variants were detected and when the patient consented, the entire exome data was analyzed. A total of 266 Dutch patients with different types of inherited eye disorders, including inherited retinal dystrophies, cataract, developmental eye disorders and optic atrophy, were investigated. In the vision gene panel analysis (likely), causative variants were detected in 49% and in the exome analysis in an additional 2% of the patients. The highest detection rate of (likely) causative variants was in patients with inherited retinal dystrophies, for instance a yield of 63% in patients with retinitis pigmentosa. In patients with developmental eye defects, cataract and optic atrophy, the detection rate was 50, 33 and 17%, respectively. An exome-sequencing approach enables a genetic diagnosis in patients with different types of inherited eye disorders using one test. The exome approach has the same detection rate as targeted panel sequencing tests, but offers a number of advantages. For instance, the vision gene panel can be frequently and easily updated with additional (novel) eye disorder genes. Determination of the genetic diagnosis improved the clinical diagnosis, regarding the assessment of the inheritance pattern as well as future disease perspective.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Finger RP, Fimmers R, Holz FG, Scholl HP: Incidence of blindness and severe visual impairment in Germany: projections for 2030. Invest Ophthalmol Vis Sci 2011; 52: 4381–4389. - PubMed
-
- Rahi JS, Dezateux C, , British Congenital Cataract Interest G: Measuring and interpreting the incidence of congenital ocular anomalies: lessons from a national study of congenital cataract in the UK. Invest Ophthalmol Vis Sci 2001; 42: 1444–1448. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

