Testicular Dnmt3 expression and global DNA methylation are down-regulated by gonadotropin releasing hormones in the ricefield eel Monopterus albus
- PMID: 28225069
- PMCID: PMC5320511
- DOI: 10.1038/srep43158
Testicular Dnmt3 expression and global DNA methylation are down-regulated by gonadotropin releasing hormones in the ricefield eel Monopterus albus
Abstract
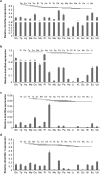
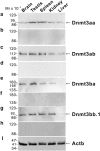
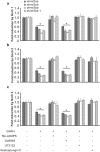
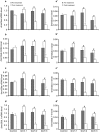
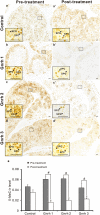
In vertebrates, DNA methyltransferase 3 (Dnmt3) homologues are responsible for de novo DNA methylation and play important roles in germ cell development. In the present study, four dnmt3 genes, dnmt3aa, dnmt3ab, dnmt3ba and dnmt3bb.1, were identified in ricefield eels. Real-time quantitative PCR analysis showed that all four dnmt3 mRNAs were detected broadly in tissues examined, with testicular expression at relatively high levels. In the testis, immunostaining for all four Dnmt3 forms was mainly localized to spermatocytes, which also contained highly methylated DNA. All three forms of Gonadotropin-releasing hormone (Gnrh) in the ricefield eel were shown to decrease the expression of dnmt3 genes in the in vitro incubated testicular fragments through cAMP and IP3/Ca2+ pathways. Moreover, in vivo treatment of male fish with three forms of Gnrh decreased significantly the testicular Dnmt3 expression at both mRNA and protein levels, and the global DNA methylation levels. These results suggest that the expression of Dnmt3 and global DNA methylation in the testis of ricefield eels are potentially down-regulated by Gnrh, and reveal a novel regulatory mechanism of testicular Dnmt3 expression in vertebrates.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
-
- Reik W., Dean W. & Walter J. Epigenetic reprogramming in mammalian development. Science 293, 1089–1093 (2001). - PubMed
-
- Goll M. G. & Bestor T. H. Eukaryotic cytosine methyltransferases. Annu Rev Biochem 74, 481–514 (2005). - PubMed
-
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev 16, 6–21 (2002). - PubMed
-
- Okano M., Bell D. W., Haber D. A. & Li E. DNA Methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257 (1999). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

