A distal intergenic region controls pancreatic endocrine differentiation by acting as a transcriptional enhancer and as a polycomb response element
- PMID: 28225770
- PMCID: PMC5321433
- DOI: 10.1371/journal.pone.0171508
A distal intergenic region controls pancreatic endocrine differentiation by acting as a transcriptional enhancer and as a polycomb response element
Abstract
Lineage-selective expression of developmental genes is dependent on the interplay between activating and repressive mechanisms. Gene activation is dependent on cell-specific transcription factors that recognize transcriptional enhancer sequences. Gene repression often depends on the recruitment of Polycomb group (PcG) proteins, although the sequences that underlie the recruitment of PcG proteins, also known as Polycomb response elements (PREs), remain poorly understood in vertebrates. While distal PREs have been identified in mammals, a role for positive-acting enhancers in PcG-mediated repression has not been described. Here we have used a highly efficient procedure based on lentiviral-mediated transgenesis to carry out in vivo fine-mapping of, cis-regulatory sequences that control lineage-specific activation of Neurog3, a master regulator of pancreatic endocrine differentiation. Our findings reveal an enhancer region that is sufficient to drive correct spacio-temporal expression of Neurog3 and demonstrate that this same region serves as a PRE in alternative lineages where Neurog3 is inactive.
Conflict of interest statement
Figures
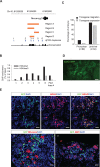

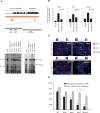
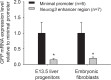
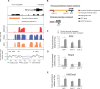
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

