Whole-genome duplication and molecular evolution in Cornus L. (Cornaceae) - Insights from transcriptome sequences
- PMID: 28225773
- PMCID: PMC5321274
- DOI: 10.1371/journal.pone.0171361
Whole-genome duplication and molecular evolution in Cornus L. (Cornaceae) - Insights from transcriptome sequences
Abstract
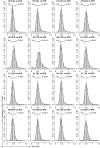
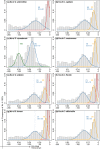
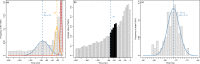
The pattern and rate of genome evolution have profound consequences in organismal evolution. Whole-genome duplication (WGD), or polyploidy, has been recognized as an important evolutionary mechanism of plant diversification. However, in non-model plants the molecular signals of genome duplications have remained largely unexplored. High-throughput transcriptome data from next-generation sequencing have set the stage for novel investigations of genome evolution using new bioinformatic and methodological tools in a phylogenetic framework. Here we compare ten de novo-assembled transcriptomes representing the major lineages of the angiosperm genus Cornus (dogwood) and relevant outgroups using a customized pipeline for analyses. Using three distinct approaches, molecular dating of orthologous genes, analyses of the distribution of synonymous substitutions between paralogous genes, and examination of substitution rates through time, we detected a shared WGD event in the late Cretaceous across all taxa sampled. The inferred doubling event coincides temporally with the paleoclimatic changes associated with the initial divergence of the genus into three major lineages. Analyses also showed an acceleration of rates of molecular evolution after WGD. The highest rates of molecular evolution were observed in the transcriptome of the herbaceous lineage, C. canadensis, a species commonly found at higher latitudes, including the Arctic. Our study demonstrates the value of transcriptome data for understanding genome evolution in closely related species. The results suggest dramatic increase in sea surface temperature in the late Cretaceous may have contributed to the evolution and diversification of flowering plants.
Conflict of interest statement
Figures

References
-
- Lynch M, Walsh B. The origins of genome architecture: Sinauer Associates Sunderland; 2007.
-
- Soltis PS, Soltis DE. Polyploidy and genome evolution: Springer; 2012.
-
- Wendel JF. Genome evolution in polyploids Plant molecular evolution: Springer; 2000. p. 225–49. - PubMed
-
- Stebbins C Jr. Variation and evolution in plants. Variation and evolution in plants. 1950.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

