Antibody Responses to Zika Virus Infections in Environments of Flavivirus Endemicity
- PMID: 28228395
- PMCID: PMC5382833
- DOI: 10.1128/CVI.00036-17
Antibody Responses to Zika Virus Infections in Environments of Flavivirus Endemicity
Abstract
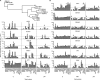
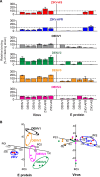
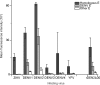
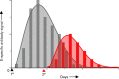
Zika virus (ZIKV) infections occur in areas where dengue virus (DENV), West Nile virus (WNV), yellow fever virus (YFV), and other viruses of the genus Flavivirus cocirculate. The envelope (E) proteins of these closely related flaviviruses induce specific long-term immunity, yet subsequent infections are associated with cross-reactive antibody responses that may enhance disease susceptibility and severity. To gain a better understanding of ZIKV infections against a background of similar viral diseases, we examined serological immune responses to ZIKV, WNV, DENV, and YFV infections of humans and nonhuman primates (NHPs). Using printed microarrays, we detected very specific antibody responses to primary infections with probes of recombinant E proteins from 15 species and lineages of flaviviruses pathogenic to humans, while high cross-reactivity between ZIKV and DENV was observed with 11 printed native viruses. Notably, antibodies from human primary ZIKV or secondary DENV infections that occurred in areas where flavivirus is endemic broadly recognized E proteins from many flaviviruses, especially DENV, indicating a strong influence of infection history on immune responses. A predictive algorithm was used to tentatively identify previous encounters with specific flaviviruses based on serum antibody interactions with the multispecies panel of E proteins. These results illustrate the potential impact of exposure to related viruses on the outcome of ZIKV infection and offer considerations for development of vaccines and diagnostics.
Keywords: Zika; cross-reactivity; flavivirus; humoral immunity; protein microarray.
Copyright © 2017 American Society for Microbiology.
Figures


References
-
- Ferreira-de-Brito A, Ribeiro IP, Miranda RM, Fernandes RS, Campos SS, Silva KA, Castro MG, Bonaldo MC, Brasil P, Lourenco-de-Oliveira R. 2016. First detection of natural infection of Aedes aegypti with Zika virus in Brazil and throughout South America. Mem Inst Oswaldo Cruz 111:655–658. doi: 10.1590/0074-02760160332. - DOI - PMC - PubMed
-
- Cauchemez S, Besnard M, Bompard P, Dub T, Guillemette-Artur P, Eyrolle-Guignot D, Salje H, Van Kerkhove MD, Abadie V, Garel C, Fontanet A, Mallet HP. 2016. Association between Zika virus and microcephaly in French Polynesia, 2013-15: a retrospective study. Lancet 387:2125–2132. doi: 10.1016/S0140-6736(16)00651-6. - DOI - PMC - PubMed
-
- Schuler-Faccini L, Ribeiro EM, Feitosa IM, Horovitz DD, Cavalcanti DP, Pessoa A, Doriqui MJ, Neri JI, Neto JM, Wanderley HY, Cernach M, El-Husny AS, Pone MV, Serao CL, Sanseverino MT, Brazilian Medical Genetics Society-Zika Embryopathy Task Force. 2016. Possible association between Zika virus infection and microcephaly - Brazil, 2015. MMWR Morb Mortal Wkly Rep 65:59–62. doi: 10.15585/mmwr.mm6503e2. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

