Novel layers of RNA polymerase III control affecting tRNA gene transcription in eukaryotes
- PMID: 28228471
- PMCID: PMC5356446
- DOI: 10.1098/rsob.170001
Novel layers of RNA polymerase III control affecting tRNA gene transcription in eukaryotes
Abstract
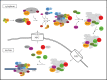
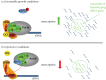
RNA polymerase III (Pol III) transcribes a limited set of short genes in eukaryotes producing abundant small RNAs, mostly tRNA. The originally defined yeast Pol III transcriptome appears to be expanding owing to the application of new methods. Also, several factors required for assembly and nuclear import of Pol III complex have been identified recently. Models of Pol III based on cryo-electron microscopy reconstructions of distinct Pol III conformations reveal unique features distinguishing Pol III from other polymerases. Novel concepts concerning Pol III functioning involve recruitment of general Pol III-specific transcription factors and distinctive mechanisms of transcription initiation, elongation and termination. Despite the short length of Pol III transcription units, mapping of transcriptionally active Pol III with nucleotide resolution has revealed strikingly uneven polymerase distribution along all genes. This may be related, at least in part, to the transcription factors bound at the internal promoter regions. Pol III uses also a specific negative regulator, Maf1, which binds to polymerase under stress conditions; however, a subset of Pol III genes is not controlled by Maf1. Among other RNA polymerases, Pol III machinery represents unique features related to a short transcript length and high transcription efficiency.
Keywords: Pol III; TFIIIC; polymerase assembly; tRNA; transcription elongation; transcription termination read-through.
© 2017 The Authors.
Figures


References
-
- Bhargava P. 2013. Epigenetic regulation of transcription by RNA polymerase III. Biochim. Biophys. Acta BBA Gene Regul. Mech. 1829, 1015–1025. (doi:10.1016/j.bbagrm.2013.05.005) - DOI - PubMed
-
- Arimbasseri AG, Maraia RJ. 2016. RNA polymerase III advances: structural and tRNA Functional Views. Trends Biochem. Sci. 41, 546–559. (doi:10.1016/j.tibs.2016.03.003) - DOI - PMC - PubMed
-
- Turowski TW, Tollervey D. 2016. Transcription by RNA polymerase III: insights into mechanism and regulation. Biochem. Soc. Trans. 44, 1367–1375. (doi:10.1042/BST20160062) - DOI - PMC - PubMed
-
- Hoffmann NA, Jakobi AJ, Moreno-Morcillo M, Glatt S, Kosinski J, Hagen WJH, Sachse C, Müller CW. 2015. Molecular structures of unbound and transcribing RNA polymerase III. Nature 528, 231–236. (doi:10.1038/nature16143) - DOI - PMC - PubMed
-
- Vannini A, Cramer P. 2012. Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Mol. Cell 45, 439–446. (doi:10.1016/j.molcel.2012.01.023) - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

