LIMCH1 regulates nonmuscle myosin-II activity and suppresses cell migration
- PMID: 28228547
- PMCID: PMC5391182
- DOI: 10.1091/mbc.E15-04-0218
LIMCH1 regulates nonmuscle myosin-II activity and suppresses cell migration
Abstract
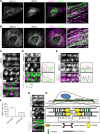
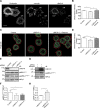
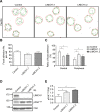
Nonmuscle myosin II (NM-II) is an important motor protein involved in cell migration. Incorporation of NM-II into actin stress fiber provides a traction force to promote actin retrograde flow and focal adhesion assembly. However, the components involved in regulation of NM-II activity are not well understood. Here we identified a novel actin stress fiber-associated protein, LIM and calponin-homology domains 1 (LIMCH1), which regulates NM-II activity. The recruitment of LIMCH1 into contractile stress fibers revealed its localization complementary to actinin-1. LIMCH1 interacted with NM-IIA, but not NM-IIB, independent of the inhibition of myosin ATPase activity with blebbistatin. Moreover, the N-terminus of LIMCH1 binds to the head region of NM-IIA. Depletion of LIMCH1 attenuated myosin regulatory light chain (MRLC) diphosphorylation in HeLa cells, which was restored by reexpression of small interfering RNA-resistant LIMCH1. In addition, LIMCH1-depleted HeLa cells exhibited a decrease in the number of actin stress fibers and focal adhesions, leading to enhanced cell migration. Collectively, our data suggest that LIMCH1 plays a positive role in regulation of NM-II activity through effects on MRLC during cell migration.
© 2017 Lin et al. This article is distributed by The American Society for Cell Biology under license from the author(s). Two months after publication it is available to the public under an Attribution–Noncommercial–Share Alike 3.0 Unported Creative Commons License (http://creativecommons.org/licenses/by-nc-sa/3.0).
Figures





References
-
- Amano M, Ito M, Kimura K, Fukata Y, Chihara K, Nakano T, Matsuura Y, Kaibuchi K. Phosphorylation and activation of myosin by Rho-associated kinase (Rho-kinase) J Biol Chem. 1996;271:20246–20249. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

