Investigation of the Role of Protein Kinase D in Human Rhinovirus Replication
- PMID: 28228588
- PMCID: PMC5391474
- DOI: 10.1128/JVI.00217-17
Investigation of the Role of Protein Kinase D in Human Rhinovirus Replication
Abstract
Picornavirus replication is known to cause extensive remodeling of Golgi and endoplasmic reticulum membranes, and a number of the host proteins involved in the viral replication complex have been identified, including oxysterol binding protein (OSBP) and phosphatidylinositol 4-kinase III beta (PI4KB). Since both OSBP and PI4KB are substrates for protein kinase D (PKD) and PKD is known to be involved in the control of Golgi membrane vesicular and lipid transport, we hypothesized that PKD played a role in viral replication. We present multiple lines of evidence in support of this hypothesis. First, infection of HeLa cells with human rhinovirus (HRV) induced the phosphorylation of PKD. Second, PKD inhibitors reduced HRV genome replication, protein expression, and titers in a concentration-dependent fashion and also blocked the replication of poliovirus (PV) and foot-and-mouth disease virus (FMDV) in a variety of cells. Third, HRV replication was significantly reduced in HeLa cells overexpressing wild-type and mutant forms of PKD1. Fourth, HRV genome replication was reduced in HAP1 cells in which the PKD1 gene was knocked out by clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9. Although we have not identified the molecular mechanism through which PKD regulates viral replication, our data suggest that this is not due to enhanced interferon signaling or an inhibition of clathrin-mediated endocytosis, and PKD inhibitors do not need to be present during viral uptake. Our data show for the first time that targeting PKD with small molecules can inhibit the replication of HRV, PV, and FMDV, and therefore, PKD may represent a novel antiviral target for drug discovery.IMPORTANCE Picornaviruses remain an important family of human and animal pathogens for which we have a very limited arsenal of antiviral agents. HRV is the causative agent of the common cold, which in itself is a relatively trivial infection; however, in asthma and chronic obstructive pulmonary disease (COPD) patients, this virus is a major cause of exacerbations resulting in an increased use of medication, worsening symptoms, and, frequently, hospital admission. Thus, HRV represents a substantial health care and economic burden for which there are no approved therapies. We sought to identify a novel host target as a potential anti-HRV therapy. HRV infection induces the phosphorylation of PKD, and inhibitors of this kinase effectively block HRV replication at an early stage of the viral life cycle. Moreover, PKD inhibitors also block PV and FMDV replication. This is the first description that PKD may represent a target for antiviral drug discovery.
Keywords: Golgi membrane; antiviral; picornavirus; protein kinase D; rhinovirus; viral replication.
Copyright © 2017 Guedán et al.
Figures



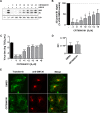

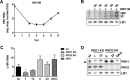
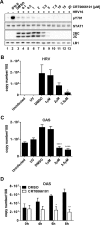

References
-
- Asher MI, Montefort S, Björkstén B, Lai CK, Strachan DP, Weiland SK, Williams H, ISAAC Phase Three Study Group. 2006. Worldwide time trends in the prevalence of symptoms of asthma, allergic rhinoconjunctivitis, and eczema in childhood: ISAAC Phases One and Three repeat multicountry cross-sectional surveys. Lancet 368:733–743. doi:10.1016/S0140-6736(06)69283-0 (Erratum, 370:1128, 2007, doi:10.1016/S0140-6736(07)61513-X.) - DOI - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- MR/K020811/1/MRC_/Medical Research Council/United Kingdom
- G1000758/MRC_/Medical Research Council/United Kingdom
- BB/L004828/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MR/M004821/1/MRC_/Medical Research Council/United Kingdom
- MC_PC_15028/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

