Drebrin-mediated microtubule-actomyosin coupling steers cerebellar granule neuron nucleokinesis and migration pathway selection
- PMID: 28230156
- PMCID: PMC5331215
- DOI: 10.1038/ncomms14484
Drebrin-mediated microtubule-actomyosin coupling steers cerebellar granule neuron nucleokinesis and migration pathway selection
Abstract
Neuronal migration from a germinal zone to a final laminar position is essential for the morphogenesis of neuronal circuits. While it is hypothesized that microtubule-actomyosin crosstalk is required for a neuron's 'two-stroke' nucleokinesis cycle, the molecular mechanisms controlling such crosstalk are not defined. By using the drebrin microtubule-actin crosslinking protein as an entry point into the cerebellar granule neuron system in combination with super-resolution microscopy, we investigate how these cytoskeletal systems interface during migration. Lattice light-sheet and structured illumination microscopy reveal a proximal leading process nanoscale architecture wherein f-actin and drebrin intervene between microtubules and the plasma membrane. Functional perturbations of drebrin demonstrate that proximal leading process microtubule-actomyosin coupling steers the direction of centrosome and somal migration, as well as the switch from tangential to radial migration. Finally, the Siah2 E3 ubiquitin ligase antagonizes drebrin function, suggesting a model for control of the microtubule-actomyosin interfaces during neuronal differentiation.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


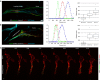
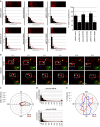

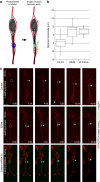


References
-
- Hatten M. E. Central nervous system neuronal migration. Annu. Rev. Neurosci. 22, 511–539 (1999). - PubMed
-
- Wynshaw-Boris A. & Gambello M. J. LIS1 and dynein motor function in neuronal migration and development. Genes Dev. 15, 639–651 (2001). - PubMed
-
- Reiner O. & Sapir T. LIS1 functions in normal development and disease. Curr. Opin. Neurobiol. 23, 951–956 (2013). - PubMed
-
- Heng J. I., Chariot A. & Nguyen L. Molecular layers underlying cytoskeletal remodelling during cortical development. Trends Neurosci. 33, 38–47 (2010). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

