Phylogenetic Characterizations of Highly Mutated EV-B106 Recombinants Showing Extensive Genetic Exchanges with Other EV-B in Xinjiang, China
- PMID: 28230168
- PMCID: PMC5322377
- DOI: 10.1038/srep43080
Phylogenetic Characterizations of Highly Mutated EV-B106 Recombinants Showing Extensive Genetic Exchanges with Other EV-B in Xinjiang, China
Abstract
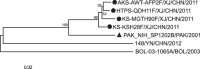
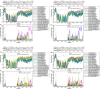
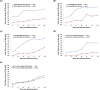
Human enterovirus B106 (EV-B106) is a new member of the enterovirus B species. To date, only three nucleotide sequences of EV-B106 have been published, and only one full-length genome sequence (the Yunnan strain 148/YN/CHN/12) is available in the GenBank database. In this study, we conducted phylogenetic characterisation of four EV-B106 strains isolated in Xinjiang, China. Pairwise comparisons of the nucleotide sequences and the deduced amino acid sequences revealed that the four Xinjiang EV-B106 strains had only 80.5-80.8% nucleotide identity and 95.4-97.3% amino acid identity with the Yunnan EV-B106 strain, indicating high mutagenicity. Similarity plots and bootscanning analyses revealed that frequent intertypic recombination occurred in all four Xinjiang EV-B106 strains in the non-structural region. These four strains may share a donor sequence with the EV-B85 strain, which circulated in Xinjiang in 2011, indicating extensive genetic exchanges between these strains. All Xinjiang EV-B106 strains were temperature-sensitive. An antibody seroprevalence study against EV-B106 in two Xinjiang prefectures also showed low titres of neutralizing antibodies, suggesting limited exposure and transmission in the population. This study contributes the whole genome sequences of EV-B106 to the GenBank database and provides valuable information regarding the molecular epidemiology of EV-B106 in China.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
-
- Picornaviridae Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses. (Elsevier, San Diego, 2011).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

