m1A Post-Transcriptional Modification in tRNAs
- PMID: 28230814
- PMCID: PMC5372732
- DOI: 10.3390/biom7010020
m1A Post-Transcriptional Modification in tRNAs
Abstract
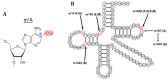
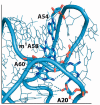
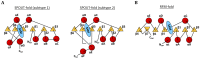
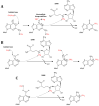
To date, about 90 post-transcriptional modifications have been reported in tRNA expanding their chemical and functional diversity. Methylation is the most frequent post-transcriptional tRNA modification that can occur on almost all nitrogen sites of the nucleobases, on the C5 atom of pyrimidines, on the C2 and C8 atoms of adenosine and, additionally, on the oxygen of the ribose 2'-OH. The methylation on the N1 atom of adenosine to form 1-methyladenosine (m1A) has been identified at nucleotide position 9, 14, 22, 57, and 58 in different tRNAs. In some cases, these modifications have been shown to increase tRNA structural stability and induce correct tRNA folding. This review provides an overview of the currently known m1A modifications, the different m1A modification sites, the biological role of each modification, and the enzyme responsible for each methylation in different species. The review further describes, in detail, two enzyme families responsible for formation of m1A at nucleotide position 9 and 58 in tRNA with a focus on the tRNA binding, m1A mechanism, protein domain organisation and overall structures.
Keywords: 1-methyladenosine; Trm10; Trm6–Trm61; TrmI; Trmt10C; m1A; tRNA, methylation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

