Biologically Relevant Heterogeneity: Metrics and Practical Insights
- PMID: 28231035
- PMCID: PMC5464733
- DOI: 10.1177/2472555216682725
Biologically Relevant Heterogeneity: Metrics and Practical Insights
Abstract
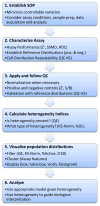
Heterogeneity is a fundamental property of biological systems at all scales that must be addressed in a wide range of biomedical applications, including basic biomedical research, drug discovery, diagnostics, and the implementation of precision medicine. There are a number of published approaches to characterizing heterogeneity in cells in vitro and in tissue sections. However, there are no generally accepted approaches for the detection and quantitation of heterogeneity that can be applied in a relatively high-throughput workflow. This review and perspective emphasizes the experimental methods that capture multiplexed cell-level data, as well as the need for standard metrics of the spatial, temporal, and population components of heterogeneity. A recommendation is made for the adoption of a set of three heterogeneity indices that can be implemented in any high-throughput workflow to optimize the decision-making process. In addition, a pairwise mutual information method is suggested as an approach to characterizing the spatial features of heterogeneity, especially in tissue-based imaging. Furthermore, metrics for temporal heterogeneity are in the early stages of development. Example studies indicate that the analysis of functional phenotypic heterogeneity can be exploited to guide decisions in the interpretation of biomedical experiments, drug discovery, diagnostics, and the design of optimal therapeutic strategies for individual patients.
Keywords: cellular models; computational pathology; drug discovery; flow cytometry; heterogeneity; high-content screening; organs-on-chips; precision medicine; quantitative systems pharmacology; systems biology.
Figures





References
-
- Krieger T, Simons BD. Dynamic stem cell heterogeneity. Development. 2015;142:1396–1406. - PubMed
-
- Gough A, Lezon T, Faeder JR, et al. High-Content Analysis with Cellular and Tissue Systems Biology: a Bridge between Cancer Cell Biology and Tissue-Based Diagnostics. In: Mendelsohn J, Howley PM, Israel MA, et al., editors. The molecular basis of cancer. 4. Chapter 25. Saunders/Elsevier; Philadelphia, PA: 2015. pp. 369–392.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

