Mindboggling morphometry of human brains
- PMID: 28231282
- PMCID: PMC5322885
- DOI: 10.1371/journal.pcbi.1005350
Mindboggling morphometry of human brains
Abstract
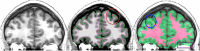
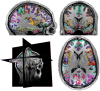
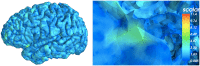
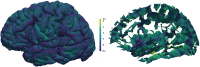
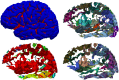
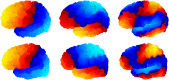
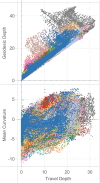
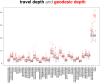
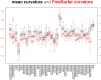
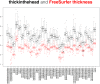
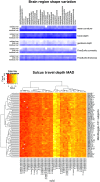
Mindboggle (http://mindboggle.info) is an open source brain morphometry platform that takes in preprocessed T1-weighted MRI data and outputs volume, surface, and tabular data containing label, feature, and shape information for further analysis. In this article, we document the software and demonstrate its use in studies of shape variation in healthy and diseased humans. The number of different shape measures and the size of the populations make this the largest and most detailed shape analysis of human brains ever conducted. Brain image morphometry shows great potential for providing much-needed biological markers for diagnosing, tracking, and predicting progression of mental health disorders. Very few software algorithms provide more than measures of volume and cortical thickness, while more subtle shape measures may provide more sensitive and specific biomarkers. Mindboggle computes a variety of (primarily surface-based) shapes: area, volume, thickness, curvature, depth, Laplace-Beltrami spectra, Zernike moments, etc. We evaluate Mindboggle's algorithms using the largest set of manually labeled, publicly available brain images in the world and compare them against state-of-the-art algorithms where they exist. All data, code, and results of these evaluations are publicly available.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Koutsouleris N, Meisenzahl EM, Davatzikos C, Bottlender R, Frodl T, Scheuerecker J, et al. Use of neuroanatomical pattern classification to identify subjects in at-risk mental states of psychosis and predict disease transition. Arch Gen Psychiatry [Internet]. 2009. July [cited 2016 Aug 6];66(7):700–12. Available from: 10.1001/archgenpsychiatry.2009.62 - DOI - PMC - PubMed
-
- Canli T, Cooney RE, Goldin P, Shah M, Sivers H, Thomason ME, et al. Amygdala reactivity to emotional faces predicts improvement in major depression. Neuroreport [Internet]. 2005. August 22;16(12):1267–70. Available from: http://www.ncbi.nlm.nih.gov/pubmed/16056122 - PubMed
-
- Chen C-H, Ridler K, Suckling J, Williams S, Fu CHY, Merlo-Pich E, et al. Brain imaging correlates of depressive symptom severity and predictors of symptom improvement after antidepressant treatment. Biol Psychiatry [Internet]. 2007. September 1 [cited 2016 Aug 6];62(5):407–14. Available from: 10.1016/j.biopsych.2006.09.018 - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

