Rapid evolution of distinct Helicobacter pylori subpopulations in the Americas
- PMID: 28231283
- PMCID: PMC5322909
- DOI: 10.1371/journal.pgen.1006546
Rapid evolution of distinct Helicobacter pylori subpopulations in the Americas
Erratum in
-
Correction: Rapid evolution of distinct Helicobacter pylori subpopulations in the Americas.PLoS Genet. 2017 Apr 14;13(4):e1006730. doi: 10.1371/journal.pgen.1006730. eCollection 2017 Apr. PLoS Genet. 2017. PMID: 28410409 Free PMC article.
Abstract
For the last 500 years, the Americas have been a melting pot both for genetically diverse humans and for the pathogenic and commensal organisms associated with them. One such organism is the stomach-dwelling bacterium Helicobacter pylori, which is highly prevalent in Latin America where it is a major current public health challenge because of its strong association with gastric cancer. By analyzing the genome sequence of H. pylori isolated in North, Central and South America, we found evidence for admixture between H. pylori of European and African origin throughout the Americas, without substantial input from pre-Columbian (hspAmerind) bacteria. In the US, strains of African and European origin have remained genetically distinct, while in Colombia and Nicaragua, bottlenecks and rampant genetic exchange amongst isolates have led to the formation of national gene pools. We found three outer membrane proteins with atypical levels of Asian ancestry in American strains, as well as alleles that were nearly fixed specifically in South American isolates, suggesting a role for the ethnic makeup of hosts in the colonization of incoming strains. Our results show that new H. pylori subpopulations can rapidly arise, spread and adapt during times of demographic flux, and suggest that differences in transmission ecology between high and low prevalence areas may substantially affect the composition of bacterial populations.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


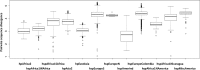
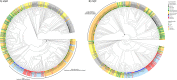


References
-
- Bianchine PJ, Russo TA (1992) The Role of Epidemic Infectious-Diseases in the Discovery of America. Allergy Proceedings 13: 225–232. - PubMed
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

