Identification of the ER-resident E3 ubiquitin ligase RNF145 as a novel LXR-regulated gene
- PMID: 28231341
- PMCID: PMC5322959
- DOI: 10.1371/journal.pone.0172721
Identification of the ER-resident E3 ubiquitin ligase RNF145 as a novel LXR-regulated gene
Abstract
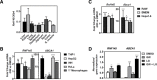
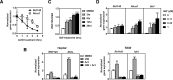
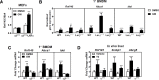
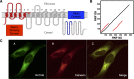
Cellular cholesterol metabolism is subject to tight regulation to maintain adequate levels of this central lipid molecule. Herein, the sterol-responsive Liver X Receptors (LXRs) play an important role owing to their ability to reduce cellular cholesterol load. In this context, identifying the full set of LXR-regulated genes will contribute to our understanding of their role in cholesterol metabolism. Using global transcriptional analysis we report here the identification of RNF145 as an LXR-regulated target gene. We demonstrate that RNF145 is regulated by LXRs in both human and mouse primary cells and cell lines, and in vivo in mice. Regulation of RNF145 by LXR depends on a functional LXR-element in its proximal promotor. Consistent with LXR-dependent regulation of Rnf145 we show that regulation is lost in macrophages and fibroblasts from Lxrαβ(-/-) mice, and also in vivo in livers of Lxrα(-/-) mice treated with the LXR synthetic ligand T0901317. RNF145 is closely related to RNF139/TRC8, an E3 ligase implicated in control of SREBP processing. However, silencing of RNF145 in HepG2 or HeLa cells does not impair SREBP1/2 processing and sterol-responsive gene expression in these cells. Similar to TRC8, we demonstrate that RNF145 is localized to the ER and that it possesses intrinsic E3 ubiquitin ligase activity. In summary, we report the identification of RNF145 as an ER-resident E3 ubiquitin ligase that is transcriptionally controlled by LXR.
Conflict of interest statement
Figures



Similar articles
-
Inhibition of cholesterol biosynthesis through RNF145-dependent ubiquitination of SCAP.Elife. 2017 Oct 25;6:e28766. doi: 10.7554/eLife.28766. Elife. 2017. PMID: 29068315 Free PMC article.
-
The sterol-responsive RNF145 E3 ubiquitin ligase mediates the degradation of HMG-CoA reductase together with gp78 and Hrd1.Elife. 2018 Dec 13;7:e40009. doi: 10.7554/eLife.40009. Elife. 2018. PMID: 30543180 Free PMC article.
-
The liver X receptors and sterol regulatory element binding proteins alter progesterone secretion and are regulated by human chorionic gonadotropin in human luteinized granulosa cells.Mol Cell Endocrinol. 2018 Sep 15;473:124-135. doi: 10.1016/j.mce.2018.01.011. Epub 2018 Jan 31. Mol Cell Endocrinol. 2018. PMID: 29366778 Free PMC article.
-
Transcriptional and posttranscriptional control of cholesterol homeostasis by liver X receptors.Cold Spring Harb Symp Quant Biol. 2011;76:129-37. doi: 10.1101/sqb.2011.76.010702. Epub 2011 Aug 22. Cold Spring Harb Symp Quant Biol. 2011. PMID: 21859674 Review.
-
Feedback regulation of cholesterol uptake by the LXR-IDOL-LDLR axis.Arterioscler Thromb Vasc Biol. 2012 Nov;32(11):2541-6. doi: 10.1161/ATVBAHA.112.250571. Epub 2012 Aug 30. Arterioscler Thromb Vasc Biol. 2012. PMID: 22936343 Free PMC article. Review.
Cited by
-
Ring finger protein 145 (RNF145) is a ubiquitin ligase for sterol-induced degradation of HMG-CoA reductase.J Biol Chem. 2018 Mar 16;293(11):4047-4055. doi: 10.1074/jbc.RA117.001260. Epub 2018 Jan 26. J Biol Chem. 2018. PMID: 29374057 Free PMC article.
-
Promoter-Specific Variants in NeuroD1 and H3K4me3 Coincident Regions and Clinical Outcomes of Small Cell Lung Cancer.J Korean Med Sci. 2023 Nov 20;38(45):e381. doi: 10.3346/jkms.2023.38.e381. J Korean Med Sci. 2023. PMID: 37987107 Free PMC article.
-
Inhibition of cholesterol biosynthesis through RNF145-dependent ubiquitination of SCAP.Elife. 2017 Oct 25;6:e28766. doi: 10.7554/eLife.28766. Elife. 2017. PMID: 29068315 Free PMC article.
-
Ubiquitination in lipid metabolism reprogramming: implications for pediatric solid tumors.Front Immunol. 2025 Apr 30;16:1554311. doi: 10.3389/fimmu.2025.1554311. eCollection 2025. Front Immunol. 2025. PMID: 40370434 Free PMC article. Review.
-
Genetic variation of RNF145 gene and blood lipid levels in Xinjiang population, China.Sci Rep. 2021 Mar 16;11(1):5969. doi: 10.1038/s41598-021-85503-z. Sci Rep. 2021. PMID: 33727652 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

