Expression and characteristics of manganese peroxidase from Ganoderma lucidum in Pichia pastoris and its application in the degradation of four dyes and phenol
- PMID: 28231778
- PMCID: PMC5324234
- DOI: 10.1186/s12896-017-0338-5
Expression and characteristics of manganese peroxidase from Ganoderma lucidum in Pichia pastoris and its application in the degradation of four dyes and phenol
Abstract
Background: Manganese peroxidase (MnP) of white rot basidiomycetes, an extracellular heme enzyme, is part of a peroxidase superfamily that is capable of degrading the different phenolic compounds. Ganoderma, a white rot basidiomycete widely distributed worldwide, could secrete lignin-modifying enzymes (LME), including laccase (Lac), lignin peroxidases (LiP) and MnP.
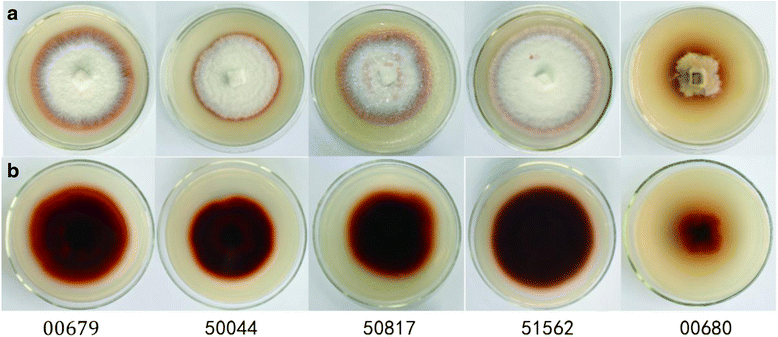
Results: After the selection of a G. lucidum strain from five Ganoderma strains, the 1092 bp full-length cDNA of the MnP gene, designated as G. lucidum MnP (GluMnP1), was cloned from the selected strain. We subsequently constructed an eukaryotic expression vector, pAO815:: GlMnP, and transferred it into Pichia pastoris SMD116. Recombinant GluMnP1 (rGluMnP1) was with a yield of 126 mg/L and a molecular weight of approximately 37.72 kDa and a specific enzyme activity of 524.61 U/L. The rGluMnP1 could be capable of the decolorization of four types of dyes and the degradation of phenol. Phenol and its principal degradation products including hydroquinone, pyrocatechol, resorcinol, benzoquinone, were detected successfully in the experiments.
Conclusions: The rGluMnP1 could be effectively expressed in Pichia pastoris and with a higher oxidation activity. We infer that, in the initial stages of the reaction, the catechol-mediated cycle should be the principal route of enzymatic degradation of phenol and its oxidation products. This study highlights the potential industrial applications associated with the production of MnP by genetic engineering methods, and the application of industrial wastewater treatment.
Keywords: Degradation; Ganoderma lucidum; Manganese peroxidase; Phenolic compound; Yeast expression system.
Figures






Similar articles
-
Heterologous expression and characterization of a dye-decolorizing peroxidase from Ganoderma lucidum, and its application in decolorization and detoxifization of different types of dyes.World J Microbiol Biotechnol. 2024 Aug 17;40(10):303. doi: 10.1007/s11274-024-04084-x. World J Microbiol Biotechnol. 2024. PMID: 39153119
-
Molecular cloning of a laccase gene from Ganoderma lucidum and heterologous expression in Pichia pastoris.J Basic Microbiol. 2014 Jul;54 Suppl 1:S134-41. doi: 10.1002/jobm.201200808. Epub 2013 May 29. J Basic Microbiol. 2014. PMID: 23720193
-
Dye decolorization and detoxification potential of Ca-alginate beads immobilized manganese peroxidase.BMC Biotechnol. 2015 Dec 10;15:111. doi: 10.1186/s12896-015-0227-8. BMC Biotechnol. 2015. PMID: 26654190 Free PMC article.
-
Harnessing the potential of white rot fungi and ligninolytic enzymes for efficient textile dye degradation: A comprehensive review.Water Environ Res. 2024 Jan;96(1):e10959. doi: 10.1002/wer.10959. Water Environ Res. 2024. PMID: 38204323 Review.
-
Enzymatic membrane reactors for biodegradation of recalcitrant compounds. Application to dye decolourisation.J Biotechnol. 2002 Nov 13;99(3):249-57. doi: 10.1016/s0168-1656(02)00217-1. J Biotechnol. 2002. PMID: 12385713 Review.
Cited by
-
Trends and Applications of Omics Technologies to Functional Characterisation of Enzymes and Protein Metabolites Produced by Fungi.J Fungi (Basel). 2021 Aug 27;7(9):700. doi: 10.3390/jof7090700. J Fungi (Basel). 2021. PMID: 34575737 Free PMC article. Review.
-
The Potential of Peroxidases Extracted from the Spent Mushroom (Flammulina velutipes) Substrate Significantly Degrade Mycotoxin Deoxynivalenol.Toxins (Basel). 2021 Jan 19;13(1):72. doi: 10.3390/toxins13010072. Toxins (Basel). 2021. PMID: 33478106 Free PMC article.
-
Challenges and Recent Advances in Enzyme-Mediated Wastewater Remediation-A Review.Nanomaterials (Basel). 2021 Nov 19;11(11):3124. doi: 10.3390/nano11113124. Nanomaterials (Basel). 2021. PMID: 34835887 Free PMC article. Review.
-
Patulin Detoxification by Recombinant Manganese Peroxidase from Moniliophthora roreri Expressed by Pichia pastoris.Toxins (Basel). 2022 Jun 29;14(7):440. doi: 10.3390/toxins14070440. Toxins (Basel). 2022. PMID: 35878178 Free PMC article.
-
Textile Dye Biodecolorization by Manganese Peroxidase: A Review.Molecules. 2021 Jul 21;26(15):4403. doi: 10.3390/molecules26154403. Molecules. 2021. PMID: 34361556 Free PMC article. Review.
References
-
- Paszczyński A, Huynh VB, Crawford R. Enzymatic activities of an extracellular, manganese-dependent peroxidase from Phanerochaete chrysosporium. FEMS Microbiol Lett. 1985;29:37–41. doi: 10.1111/j.1574-6968.1985.tb00831.x. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

