Genomic analysis of 63,220 tumors reveals insights into tumor uniqueness and targeted cancer immunotherapy strategies
- PMID: 28231819
- PMCID: PMC5324279
- DOI: 10.1186/s13073-017-0408-2
Genomic analysis of 63,220 tumors reveals insights into tumor uniqueness and targeted cancer immunotherapy strategies
Abstract
Background: The integration of genomics with immunotherapy has potential value for cancer vaccine development. Given the clinical successes of immune checkpoint modulators, interest in cancer vaccines as therapeutic options has been revived. Current data suggest that each tumor contains a unique set of mutations (mutanome), thus requiring the creation of individualized cancer vaccines. However, rigorous analysis of non-individualized cancer immunotherapy approaches across multiple cancer types and in the context of known driver alterations has yet to be reported. We therefore set out to determine the feasibility of a generalizable cancer vaccine strategy based on targeting multiple neoantigens in an HLA-A/B subtype-directed manner.
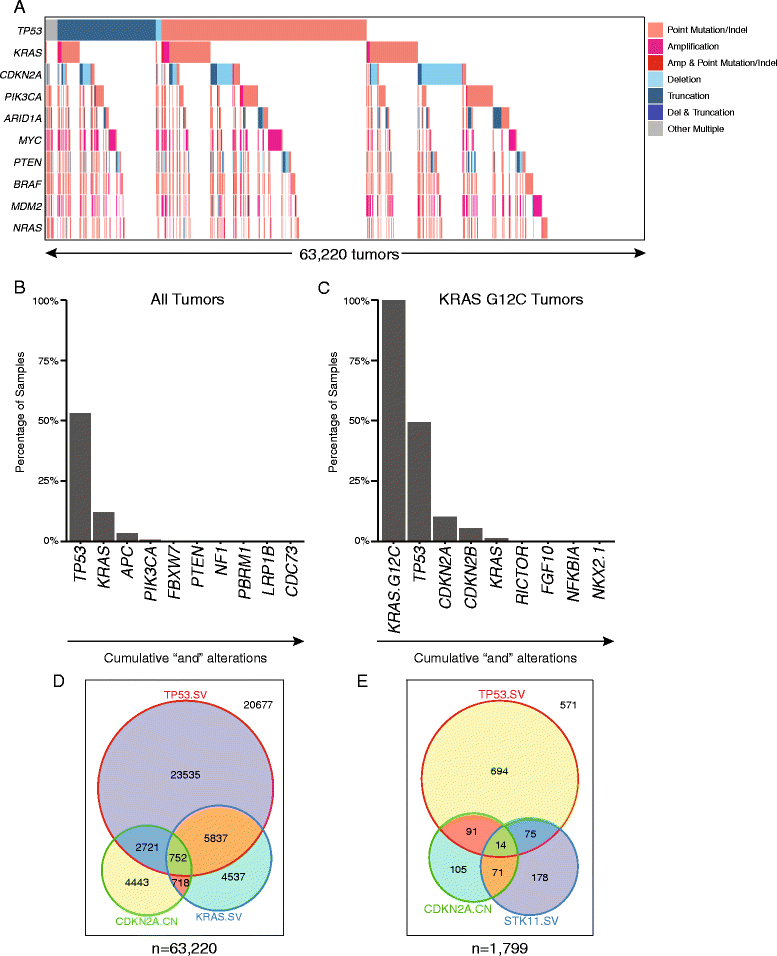
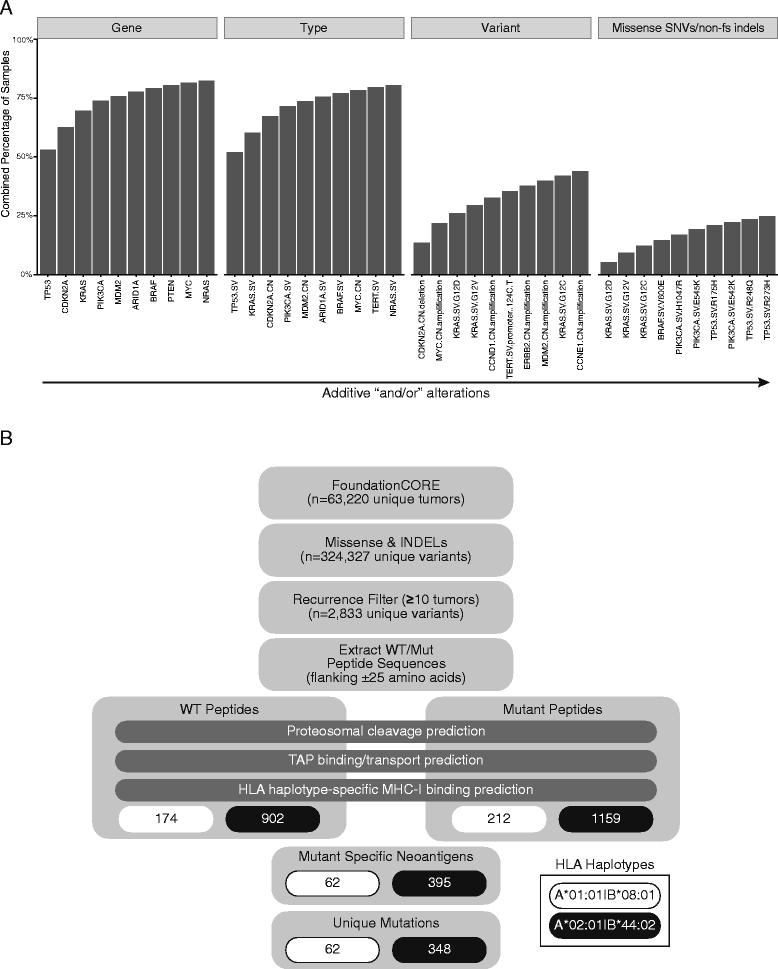
Methods: A cancer gene-focused, hybrid capture-based genomic analysis was performed on 63,220 unique tumors. Neoantigens were predicted using a combined peptide processing and MHC-I binding prediction tool (IEDB) for all recurrent (>10 tumors) missense alterations and non-frameshift indels for the two most common HLA-A/B subtypes in North American/European populations.
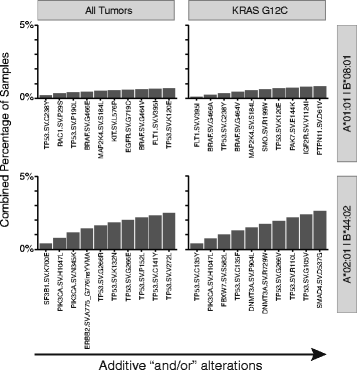
Results: Despite being overwhelmingly unique overall, many mutanomes (~45%) contain at least one mutation from a set of ten mutations chosen to maximize the number of unique tumors. This held true for tumors driven by KRAS G12C (n = 1799), PIK3CA E545K (n = 1713), or EGFR L858R (n = 478) alterations, which define distinct sample subsets. We therefore hypothesized that sets of carefully selected mutations/neoantigens may allow the development of broadly applicable semi-universal cancer vaccines. To test the feasibility of such an approach, antigen processing and MHC-I binding prediction was applied for HLA subtypes A*01:01/B*08:01 and A*02:01/B*44:02. In tumors with a specific HLA type, 0.7 and 2.5% harbored at least one of a set of ten neoantigens predicted to bind to each subtype, respectively. In comparison, KRAS G12C-driven tumors produced similar results (0.8 and 2.6% for each HLA subtype, respectively), indicating that neoantigen targets still remain highly diverse even within the context of major driver mutations.
Conclusions: This "best case scenario" analysis of a large tumor set across multiple cancer types and in the context of driver alterations reveals that semi-universal, HLA-specific cancer vaccine strategies will be relevant to only a small subset of the general population. Similar analysis of whole exome/genome sequencing, although not currently feasible at scale in a clinical setting, will likely uncover further diversity.
Keywords: Cancer vaccines; Genomic profiling; Neoantigens; Poly-epitope.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

