Epigenetic modifications and glucocorticoid sensitivity in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS)
- PMID: 28231836
- PMCID: PMC5324230
- DOI: 10.1186/s12920-017-0248-3
Epigenetic modifications and glucocorticoid sensitivity in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS)
Abstract
Background: Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS) is a debilitating idiopathic disease characterized by unexplained fatigue that fails to resolve with sufficient rest. Diagnosis is based on a list of symptoms and exclusion of other fatigue-related health conditions. Despite a heterogeneous patient population, immune and hypothalamic-pituitary-adrenal (HPA) axis function differences, such as enhanced negative feedback to glucocorticoids, are recurring findings in ME/CFS studies. Epigenetic modifications, such as CpG methylation, are known to regulate long-term phenotypic differences and previous work by our group found DNA methylome differences in ME/CFS, however the relationship between DNA methylome modifications, clinical and functional characteristics associated with ME/CFS has not been examined.
Methods: We examined the DNA methylome in peripheral blood mononuclear cells (PBMCs) of a larger cohort of female ME/CFS patients using the Illumina HumanMethylation450 BeadChip Array. In parallel to the DNA methylome analysis, we investigated in vitro glucocorticoid sensitivity differences by stimulating PBMCs with phytohaemagglutinin and suppressed growth with dexamethasone. We explored DNA methylation differences using bisulfite pyrosequencing and statistical permutation. Linear regression was implemented to discover epigenomic regions associated with self-reported quality of life and network analysis of gene ontology terms to biologically contextualize results.
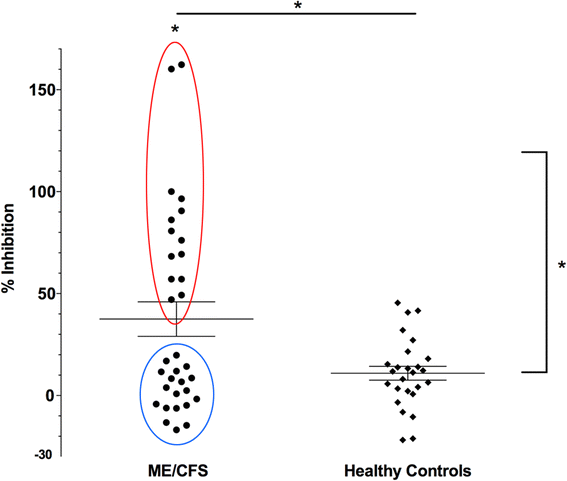
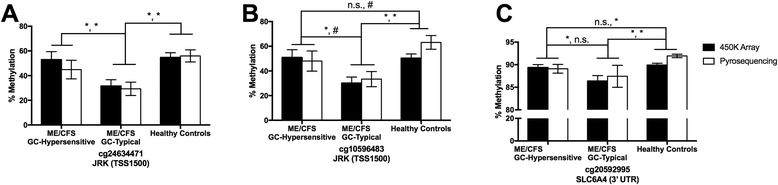
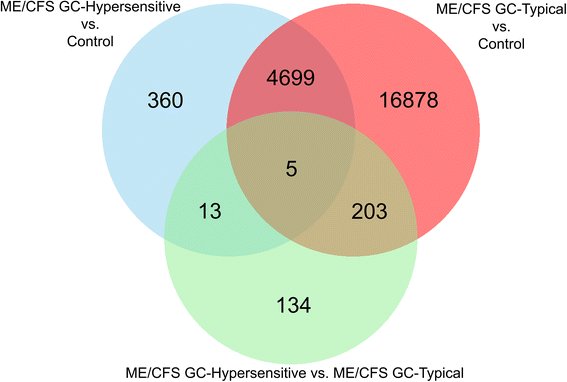
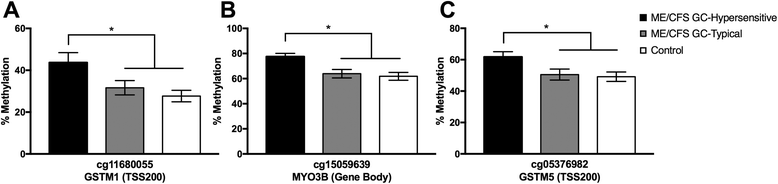
Results: We detected 12,608 differentially methylated sites between ME/CFS patients and healthy controls predominantly localized to cellular metabolism genes, some of which were also related to self-reported quality of life health scores. Among ME/CFS patients, glucocorticoid sensitivity was associated with differential methylation at 13 loci.
Conclusions: Our results indicate DNA methylation modifications in cellular metabolism in ME/CFS despite a heterogeneous patient population, implicating these processes in immune and HPA axis dysfunction in ME/CFS. Modifications to epigenetic loci associated with differences in glucocorticoid sensitivity may be important as biomarkers for future clinical testing. Overall, these findings align with recent ME/CFS work that point towards impairment in cellular energy production in this patient population.
Keywords: Chronic fatigue syndrome; Dna methylation; Epigenetics; Glucocorticoid; Hpa axis; Immune cells; Myalgic encephalomyelitis.
Figures
Similar articles
-
Identification of Myalgic Encephalomyelitis/Chronic Fatigue Syndrome-associated DNA methylation patterns.PLoS One. 2018 Jul 23;13(7):e0201066. doi: 10.1371/journal.pone.0201066. eCollection 2018. PLoS One. 2018. PMID: 30036399 Free PMC article.
-
Changes in DNA methylation profiles of myalgic encephalomyelitis/chronic fatigue syndrome patients reflect systemic dysfunctions.Clin Epigenetics. 2020 Nov 4;12(1):167. doi: 10.1186/s13148-020-00960-z. Clin Epigenetics. 2020. PMID: 33148325 Free PMC article.
-
Genome-epigenome interactions associated with Myalgic Encephalomyelitis/Chronic Fatigue Syndrome.Epigenetics. 2018;13(12):1174-1190. doi: 10.1080/15592294.2018.1549769. Epub 2018 Dec 5. Epigenetics. 2018. PMID: 30516085 Free PMC article.
-
Hypothalamic-Pituitary-Adrenal Hypofunction in Myalgic Encephalomyelitis (ME)/Chronic Fatigue Syndrome (CFS) as a Consequence of Activated Immune-Inflammatory and Oxidative and Nitrosative Pathways.Mol Neurobiol. 2017 Nov;54(9):6806-6819. doi: 10.1007/s12035-016-0170-2. Epub 2016 Oct 20. Mol Neurobiol. 2017. PMID: 27766535 Review.
-
Epigenetic Components of Myalgic Encephalomyelitis/Chronic Fatigue Syndrome Uncover Potential Transposable Element Activation.Clin Ther. 2019 Apr;41(4):675-698. doi: 10.1016/j.clinthera.2019.02.012. Epub 2019 Mar 23. Clin Ther. 2019. PMID: 30910331
Cited by
-
Behavioural modification interventions for medically unexplained symptoms in primary care: systematic reviews and economic evaluation.Health Technol Assess. 2020 Sep;24(46):1-490. doi: 10.3310/hta24460. Health Technol Assess. 2020. PMID: 32975190 Free PMC article.
-
Stress-Related Chronic Fatigue Syndrome: A Case Report with a Positive Response to Alpha-Methyl-P-Tyrosine (AMPT) Treatment.Int J Mol Sci. 2024 Jul 16;25(14):7778. doi: 10.3390/ijms25147778. Int J Mol Sci. 2024. PMID: 39063020 Free PMC article.
-
Identification of Myalgic Encephalomyelitis/Chronic Fatigue Syndrome-associated DNA methylation patterns.PLoS One. 2018 Jul 23;13(7):e0201066. doi: 10.1371/journal.pone.0201066. eCollection 2018. PLoS One. 2018. PMID: 30036399 Free PMC article.
-
Selenium-associated DNA methylation modifications in placenta and neurobehavioral development of newborns: An epigenome-wide study of two U.S. birth cohorts.Environ Int. 2020 Apr;137:105508. doi: 10.1016/j.envint.2020.105508. Epub 2020 Jan 31. Environ Int. 2020. PMID: 32007686 Free PMC article.
-
Comprehensive Circulatory Metabolomics in ME/CFS Reveals Disrupted Metabolism of Acyl Lipids and Steroids.Metabolites. 2020 Jan 14;10(1):34. doi: 10.3390/metabo10010034. Metabolites. 2020. PMID: 31947545 Free PMC article.
References
-
- Fukuda K, Straus SE, Hickie I, Sharpe MC, Dobbins JG, Komaroff A. The chronic fatigue syndrome: a comprehensive approach to its definition and study. International chronic fatigue syndrome study group. Ann Intern Med. 1994;121(12):953–959. doi: 10.7326/0003-4819-121-12-199412150-00009. - DOI - PubMed
-
- Visser J, Lentjes E, Haspels I, Graffelman W, Blauw B, de Kloet R, Nagelkerken L. Increased sensitivity to glucocorticoids in peripheral blood mononuclear cells of chronic fatigue syndrome patients, without evidence for altered density or affinity of glucocorticoid receptors. J Investig Med. 2001;49(2):195–204. doi: 10.2310/6650.2001.34047. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

