Estimation of flux ratios without uptake or release data: Application to serine and methionine metabolism
- PMID: 28232235
- PMCID: PMC5563485
- DOI: 10.1016/j.ymben.2017.02.005
Estimation of flux ratios without uptake or release data: Application to serine and methionine metabolism
Abstract
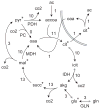
Model-based metabolic flux analysis (MFA) using isotope-labeled substrates has provided great insight into intracellular metabolic activities across a host of organisms. One challenge with applying MFA in mammalian systems, however, is the need for absolute quantification of nutrient uptake, biomass composition, and byproduct release fluxes. Such measurements are often not feasible in complex culture systems or in vivo. One way to address this issue is to estimate flux ratios, the fractional contribution of a flux to a metabolite pool, which are independent of absolute measurements and yet informative for cellular metabolism. Prior work has focused on "local" estimation of a handful of flux ratios for specific metabolites and reactions. Here, we perform systematic, model-based estimation of all flux ratios in a metabolic network using isotope labeling data, in the absence of uptake/release data. In a series of examples, we investigate what flux ratios can be well estimated with reasonably tight confidence intervals, and contrast this with confidence intervals on normalized fluxes. We find that flux ratios can provide useful information on the metabolic state, and is complementary to normalized fluxes: for certain metabolic reactions, only flux ratios can be well estimated, while for others normalized fluxes can be obtained. Simulation studies of a large human metabolic network model suggest that estimation of flux ratios is technically feasible for complex networks, but additional studies on data from actual isotopomer labeling experiments are needed to validate these results. Finally, we experimentally study serine and methionine metabolism in cancer cells using flux ratios. We find that, in these cells, the methionine cycle is truncated with little remethylation from homocysteine, and polyamine synthesis in the absence of methionine salvage leads to loss of 5-methylthioadenosine, suggesting a new mode of overflow metabolism in cancer cells. This work highlights the potential for flux ratio analysis in the absence of absolute quantification, which we anticipate will be important for both in vitro and in vivo studies of cancer metabolism.
Keywords: Cancer metabolism; Metabolic flux analysis; Metabolic network; Methionine; Optimization; Serine; Simulation studies.
Copyright © 2017 International Metabolic Engineering Society. Published by Elsevier Inc. All rights reserved.
Figures
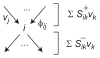




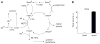
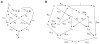
Similar articles
-
Systematic comparison of local approaches for isotopically nonstationary metabolic flux analysis.Front Plant Sci. 2023 Jun 6;14:1178239. doi: 10.3389/fpls.2023.1178239. eCollection 2023. Front Plant Sci. 2023. PMID: 37346134 Free PMC article.
-
Efficient Modeling of MS/MS Data for Metabolic Flux Analysis.PLoS One. 2015 Jul 31;10(7):e0130213. doi: 10.1371/journal.pone.0130213. eCollection 2015. PLoS One. 2015. PMID: 26230524 Free PMC article.
-
In Silico Prediction of Metabolic Fluxes in Cancer Cells with Altered S-adenosylmethionine Decarboxylase Activity.Cell Biochem Biophys. 2021 Mar;79(1):37-48. doi: 10.1007/s12013-020-00949-8. Epub 2020 Oct 11. Cell Biochem Biophys. 2021. PMID: 33040301
-
Designer labels for plant metabolism: statistical design of isotope labeling experiments for improved quantification of flux in complex plant metabolic networks.Mol Biosyst. 2013 Jan 27;9(1):99-112. doi: 10.1039/c2mb25253h. Epub 2012 Nov 1. Mol Biosyst. 2013. PMID: 23114423
-
Metabolic Flux Analysis-Linking Isotope Labeling and Metabolic Fluxes.Metabolites. 2020 Nov 6;10(11):447. doi: 10.3390/metabo10110447. Metabolites. 2020. PMID: 33172051 Free PMC article. Review.
Cited by
-
CYP2C19 and 3A4 Dominate Metabolic Clearance and Bioactivation of Terbinafine Based on Computational and Experimental Approaches.Chem Res Toxicol. 2019 Jun 17;32(6):1151-1164. doi: 10.1021/acs.chemrestox.9b00006. Epub 2019 Apr 10. Chem Res Toxicol. 2019. PMID: 30925039 Free PMC article.
-
Global 13C tracing and metabolic flux analysis of intact human liver tissue ex vivo.Nat Metab. 2024 Oct;6(10):1963-1975. doi: 10.1038/s42255-024-01119-3. Epub 2024 Aug 29. Nat Metab. 2024. PMID: 39210089 Free PMC article.
-
Isotope tracing-based metabolite identification for mass spectrometry metabolomics.bioRxiv [Preprint]. 2025 Apr 8:2025.04.07.647691. doi: 10.1101/2025.04.07.647691. bioRxiv. 2025. PMID: 40291727 Free PMC article. Preprint.
References
-
- Antoniewicz MR, Kelleher JK, Stephanopoulos G. Determination of confidence intervals of metabolic fluxes estimated from stable isotope measurements. Metabolic engineering. 2006 Jul;8(4):324–37. - PubMed
-
- Cavuoto P, Fenech MF. A review of methionine dependency and the role of methionine restriction in cancer growth control and life-span extension. Cancer treatment reviews. 2012;38(6):726–736. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

