Direct evidence that an extended hydrogen-bonding network influences activation of pyridoxal 5'-phosphate in aspartate aminotransferase
- PMID: 28232482
- PMCID: PMC5392587
- DOI: 10.1074/jbc.M116.774588
Direct evidence that an extended hydrogen-bonding network influences activation of pyridoxal 5'-phosphate in aspartate aminotransferase
Abstract
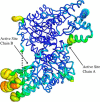
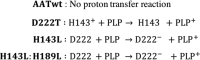
Pyridoxal 5'-phosphate (PLP) is a fundamental, multifunctional enzyme cofactor used to catalyze a wide variety of chemical reactions involved in amino acid metabolism. PLP-dependent enzymes optimize specific chemical reactions by modulating the electronic states of PLP through distinct active site environments. In aspartate aminotransferase (AAT), an extended hydrogen bond network is coupled to the pyridinyl nitrogen of the PLP, influencing the electrophilicity of the cofactor. This network, which involves residues Asp-222, His-143, Thr-139, His-189, and structural waters, is located at the edge of PLP opposite the reactive Schiff base. We demonstrate that this hydrogen bond network directly influences the protonation state of the pyridine nitrogen of PLP, which affects the rates of catalysis. We analyzed perturbations caused by single- and double-mutant variants using steady-state kinetics, high resolution X-ray crystallography, and quantum chemical calculations. Protonation of the pyridinyl nitrogen to form a pyridinium cation induces electronic delocalization in the PLP, which correlates with the enhancement in catalytic rate in AAT. Thus, PLP activation is controlled by the proximity of the pyridinyl nitrogen to the hydrogen bond microenvironment. Quantum chemical calculations indicate that Asp-222, which is directly coupled to the pyridinyl nitrogen, increases the pKa of the pyridine nitrogen and stabilizes the pyridinium cation. His-143 and His-189 also increase the pKa of the pyridine nitrogen but, more significantly, influence the position of the proton that resides between Asp-222 and the pyridinyl nitrogen. These findings indicate that the second shell residues directly enhance the rate of catalysis in AAT.
Keywords: X-ray crystallography; enzyme catalysis; hydrogen bond; pyridoxal phosphate; quantum chemistry.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures

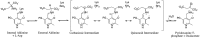




Similar articles
-
Light-enhanced catalysis by pyridoxal phosphate-dependent aspartate aminotransferase.J Am Chem Soc. 2010 Dec 1;132(47):16953-61. doi: 10.1021/ja107054x. Epub 2010 Nov 8. J Am Chem Soc. 2010. PMID: 21058708 Free PMC article.
-
NMR studies of protonation and hydrogen bond states of internal aldimines of pyridoxal 5'-phosphate acid-base in alanine racemase, aspartate aminotransferase, and poly-L-lysine.J Am Chem Soc. 2013 Dec 4;135(48):18160-75. doi: 10.1021/ja408988z. Epub 2013 Nov 21. J Am Chem Soc. 2013. PMID: 24147985
-
Strain is more important than electrostatic interaction in controlling the pKa of the catalytic group in aspartate aminotransferase.Biochemistry. 2001 Jan 16;40(2):353-60. doi: 10.1021/bi001403e. Biochemistry. 2001. PMID: 11148029
-
Aspartate aminotransferase: an old dog teaches new tricks.Arch Biochem Biophys. 2014 Feb 15;544:119-27. doi: 10.1016/j.abb.2013.10.002. Epub 2013 Oct 9. Arch Biochem Biophys. 2014. PMID: 24121043 Free PMC article. Review.
-
Critical hydrogen bonds and protonation states of pyridoxal 5'-phosphate revealed by NMR.Biochim Biophys Acta. 2011 Nov;1814(11):1426-37. doi: 10.1016/j.bbapap.2011.06.004. Epub 2011 Jun 16. Biochim Biophys Acta. 2011. PMID: 21703367 Review.
Cited by
-
Biochemical Characterization and Structure-Based Mutational Analysis Provide Insight into the Binding and Mechanism of Action of Novel Aspartate Aminotransferase Inhibitors.Biochemistry. 2018 Nov 27;57(47):6604-6614. doi: 10.1021/acs.biochem.8b00914. Epub 2018 Nov 12. Biochemistry. 2018. PMID: 30365304 Free PMC article.
-
Saccharomyces cerevisiae Differential Functionalization of Presumed ScALT1 and ScALT2 Alanine Transaminases Has Been Driven by Diversification of Pyridoxal Phosphate Interactions.Front Microbiol. 2018 May 14;9:944. doi: 10.3389/fmicb.2018.00944. eCollection 2018. Front Microbiol. 2018. PMID: 29867852 Free PMC article.
-
Discovery of GOT1 Inhibitors from a Marine-Derived Aspergillus terreus That Act against Pancreatic Ductal Adenocarcinoma.Mar Drugs. 2021 Oct 20;19(11):588. doi: 10.3390/md19110588. Mar Drugs. 2021. PMID: 34822459 Free PMC article.
-
Structure of 3-mercaptopropionic acid dioxygenase with a substrate analog reveals bidentate substrate binding at the iron center.J Biol Chem. 2021 Jan-Jun;296:100492. doi: 10.1016/j.jbc.2021.100492. Epub 2021 Mar 1. J Biol Chem. 2021. PMID: 33662397 Free PMC article.
-
Direct visualization of critical hydrogen atoms in a pyridoxal 5'-phosphate enzyme.Nat Commun. 2017 Oct 16;8(1):955. doi: 10.1038/s41467-017-01060-y. Nat Commun. 2017. PMID: 29038582 Free PMC article.
References
-
- Toney M. D. (2005) Reaction specificity in pyridoxal phosphate enzymes. Arch. Biochem. Biophys. 433, 279–287 - PubMed
-
- Oliveira E. F., Cerqueira N. M., Fernandes P. A., and Ramos M. J. (2011) Mechanism of formation of the internal aldimine in pyridoxal 5′-phosphate-dependent enzymes. J. Am. Chem. Soc. 133, 15496–15505 - PubMed
-
- Hayashi H., and Kagamiyama H. (1997) Transient-state kinetics of the reaction of aspartate aminotransferase with aspartate at low pH reveals dual routes in the enzyme-substrate association process. Biochemistry 36, 13558–13569 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

