Evolutionary Conservation of ABA Signaling for Stomatal Closure
- PMID: 28232585
- PMCID: PMC5462018
- DOI: 10.1104/pp.16.01848
Evolutionary Conservation of ABA Signaling for Stomatal Closure
Abstract
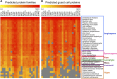
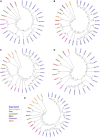
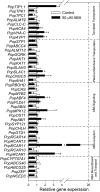
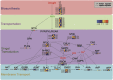
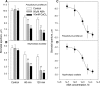
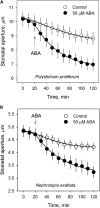
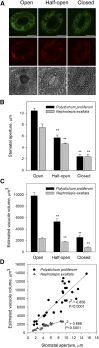
Abscisic acid (ABA)-driven stomatal regulation reportedly evolved after the divergence of ferns, during the early evolution of seed plants approximately 360 million years ago. This hypothesis is based on the observation that the stomata of certain fern species are unresponsive to ABA, but exhibit passive hydraulic control. However, ABA-induced stomatal closure was detected in some mosses and lycophytes. Here, we observed that a number of ABA signaling and membrane transporter protein families diversified over the evolutionary history of land plants. The aquatic ferns Azolla filiculoides and Salvinia cucullata have representatives of 23 families of proteins orthologous to those of Arabidopsis (Arabidopsis thaliana) and all other land plant species studied. Phylogenetic analysis of the key ABA signaling proteins indicates an evolutionarily conserved stomatal response to ABA. Moreover, comparative transcriptomic analysis has identified a suite of ABA-responsive genes that differentially expressed in a terrestrial fern species, Polystichum proliferum These genes encode proteins associated with ABA biosynthesis, transport, reception, transcription, signaling, and ion and sugar transport, which fit the general ABA signaling pathway constructed from Arabidopsis and Hordeum vulgare The retention of these key ABA-responsive genes could have had a profound effect on the adaptation of ferns to dry conditions. Furthermore, stomatal assays have shown the primary evidence for ABA-induced closure of stomata in two terrestrial fern species Pproliferum and Nephrolepis exaltata In summary, we report, to our knowledge, new molecular and physiological evidence for the presence of active stomatal control in ferns.
© 2017 American Society of Plant Biologists. All Rights Reserved.
Figures

References
-
- Beerling DJ, Franks PJ (2009) Evolution of stomatal function in ‘lower’ land plants. New Phytol 183: 921–925 - PubMed
-
- Berry JA, Beerling DJ, Franks PJ (2010) Stomata: key players in the earth system, past and present. Curr Opin Plant Biol 13: 233–240 - PubMed
-
- Blatt MR. (2000) Cellular signaling and volume control in stomatal movements in plants. Annu Rev Cell Dev Biol 16: 221–241 - PubMed
-
- Blatt MR, Armstrong F (1993) K+ channels of stomatal guard cells: abscisic-acid-evoked control of the outward rectifier mediated by cytoplasmic pH. Planta 191: 330–341
-
- Blatt MR, Thiel G, Trentham DR (1990) Reversible inactivation of K+ channels of Vicia stomatal guard cells following the photolysis of caged inositol 1,4,5-trisphosphate. Nature 346: 766–769 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

