Unreported intrinsic disorder in proteins: Building connections to the literature on IDPs
- PMID: 28232880
- PMCID: PMC5314882
- DOI: 10.4161/21690693.2014.970499
Unreported intrinsic disorder in proteins: Building connections to the literature on IDPs
Abstract
This review opens a new series entitled "Unreported intrinsic disorder in proteins." The goal of this series is to bring attention of researchers to an interesting phenomenon of missed (or overlooked, or ignored, or unreported) disorder. This series serves as a companion to "Digested Disorder" which provides a quarterly review of papers on intrinsically disordered proteins (IDPs) found by standard literature searches. The need for this alternative series results from the observation that there are numerous publications that describe IDPs (or hybrid proteins with ordered and disordered regions) yet fail to recognize many of the key discoveries and publications in the IDP field. By ignoring the body of work on IDPs, such publications often fail to relate their findings to prior discoveries or fail to explore the obvious implications of their work. Thus, the goal of this series is not only to review these very interesting and important papers, but also to point out how each paper relates to the IDP field and show how common tools in the IDP field can readily take the findings in new directions or provide a broader context for the reported findings.
Keywords: intrinsically disordered protein; molecular recognition; posttranslational modification; protein function; protein-protein interaction.
Figures


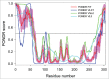
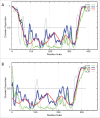
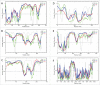
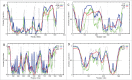




References
-
- Uversky VN. Digested disorder: quarterly intrinsic disorder digest (January/February/ March, 2013). Intrinsically Disord Proteins 2013; 1:e25496; http://dx.doi.org/ 10.4161/idp.25496 - DOI - PMC - PubMed
-
- DeForte S, Reddy KD, Uversky VN. Digested disorder, issue #2: quarterly intrinsic disorder digest (April/May/June, 2013). Intrinsically Disord Prot 2013; 1:e27454; http://dx.doi.org/ 10.4161/idp.27454 - DOI - PMC - PubMed
-
- Reddy KD, DeForte S, Uversky VN. Digested disorder, issue #3: quarterly intrinsic disorder digest (July-August-September, 2013). Intrinsically Disord Prot 2014; 2:e27833; http://dx.doi.org/ 10.4161/idp.27833 - DOI - PMC - PubMed
-
- Peng ZL, Kurgan L. Comprehensive comparative assessment of in-silico predictors of disordered regions. Curr Protein Pept Sci 2012; 13:6-18; PMID:22044149; http://dx.doi.org/ 10.2174/138920312799277938 - DOI - PubMed
-
- Andreou AZ, Klostermeier D. eIF4B and eIF4G jointly stimulate eIF4A ATPase and unwinding activities by modulation of the eIF4A conformational cycle. J Mol Biol 2014; 426:51-61; PMID:24080224; http://dx.doi.org/ 10.1016/j.jmb.2013.09.027 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
