Caulobacter crescentus CdnL is a non-essential RNA polymerase-binding protein whose depletion impairs normal growth and rRNA transcription
- PMID: 28233804
- PMCID: PMC5324124
- DOI: 10.1038/srep43240
Caulobacter crescentus CdnL is a non-essential RNA polymerase-binding protein whose depletion impairs normal growth and rRNA transcription
Abstract
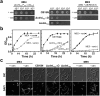
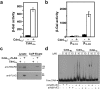
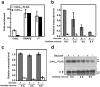
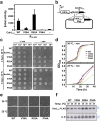
CdnL is an essential RNA polymerase (RNAP)-binding activator of rRNA transcription in mycobacteria and myxobacteria but reportedly not in Bacillus. Whether its function and mode of action are conserved in other bacteria thus remains unclear. Because virtually all alphaproteobacteria have a CdnL homolog and none of these have been characterized, we studied the homolog (CdnLCc) of the model alphaproteobacterium Caulobacter crescentus. We show that CdnLCc is not essential for viability but that its absence or depletion causes slow growth and cell filamentation. CdnLCc is degraded in vivo in a manner dependent on its C-terminus, yet excess CdnLCc resulting from its stabilization did not adversely affect growth. We find that CdnLCc interacts with itself and with the RNAP β subunit, and localizes to at least one rRNA promoter in vivo, whose activity diminishes upon depletion of CdnLCc. Interestingly, cells expressing CdnLCc mutants unable to interact with the RNAP were cold-sensitive, suggesting that CdnLCc interaction with RNAP is especially required at lower than standard growth temperatures in C. crescentus. Our study indicates that despite limited sequence similarities and regulatory differences compared to its myco/myxobacterial homologs, CdnLCc may share similar biological functions, since it affects rRNA synthesis, probably by stabilizing open promoter-RNAP complexes.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

Similar articles
-
Structure-function dissection of Myxococcus xanthus CarD N-terminal domain, a defining member of the CarD_CdnL_TRCF family of RNA polymerase interacting proteins.PLoS One. 2015 Mar 26;10(3):e0121322. doi: 10.1371/journal.pone.0121322. eCollection 2015. PLoS One. 2015. PMID: 25811865 Free PMC article.
-
Effects of Increasing the Affinity of CarD for RNA Polymerase on Mycobacterium tuberculosis Growth, rRNA Transcription, and Virulence.J Bacteriol. 2017 Jan 30;199(4):e00698-16. doi: 10.1128/JB.00698-16. Print 2017 Feb 15. J Bacteriol. 2017. PMID: 27920294 Free PMC article.
-
Structural insights into RNA polymerase recognition and essential function of Myxococcus xanthus CdnL.PLoS One. 2014 Oct 1;9(10):e108946. doi: 10.1371/journal.pone.0108946. eCollection 2014. PLoS One. 2014. PMID: 25272012 Free PMC article.
-
RNA-controlled regulation in Caulobacter crescentus.Curr Opin Microbiol. 2021 Apr;60:1-7. doi: 10.1016/j.mib.2021.01.002. Epub 2021 Jan 30. Curr Opin Microbiol. 2021. PMID: 33529919 Review.
-
Cis- and trans-acting elements required for regulation of flagellar gene transcription in the bacterium Caulobacter crescentus.Cell Mol Biol Res. 1993;39(4):361-9. Cell Mol Biol Res. 1993. PMID: 8312972 Review.
Cited by
-
Clp protease and antisense RNA jointly regulate the global regulator CarD to mediate mycobacterial starvation response.Elife. 2022 Jan 26;11:e73347. doi: 10.7554/eLife.73347. Elife. 2022. PMID: 35080493 Free PMC article.
-
The DUF1013 protein TrcR tracks with RNA polymerase to control the bacterial cell cycle and protect against antibiotics.Proc Natl Acad Sci U S A. 2021 Feb 23;118(8):e2010357118. doi: 10.1073/pnas.2010357118. Proc Natl Acad Sci U S A. 2021. PMID: 33602809 Free PMC article.
-
Plasticity in oligomerization, operator architecture, and DNA binding in the mode of action of a bacterial B12-based photoreceptor.J Biol Chem. 2018 Nov 16;293(46):17888-17905. doi: 10.1074/jbc.RA118.004838. Epub 2018 Sep 27. J Biol Chem. 2018. PMID: 30262667 Free PMC article.
-
The conserved transcriptional regulator CdnL is required for metabolic homeostasis and morphogenesis in Caulobacter.PLoS Genet. 2020 Jan 21;16(1):e1008591. doi: 10.1371/journal.pgen.1008591. eCollection 2020 Jan. PLoS Genet. 2020. PMID: 31961855 Free PMC article.
-
A majority of Rhodobacter sphaeroides promoters lack a crucial RNA polymerase recognition feature, enabling coordinated transcription activation.Proc Natl Acad Sci U S A. 2020 Nov 24;117(47):29658-29668. doi: 10.1073/pnas.2010087117. Epub 2020 Nov 9. Proc Natl Acad Sci U S A. 2020. PMID: 33168725 Free PMC article.
References
-
- Lee D. J., Minchin S. D. & Busby S. J. Activating transcription in bacteria. Annu. Rev. Microbiol. 66, 125–152 (2012). - PubMed
-
- Abellón-Ruiz J. et al. The CarD/CarG regulatory complex is required for the action of several members of the large set of Myxococcus xanthus extracytoplasmic function sigma factors. Environ. Microbiol. 16, 2475–2490 (2014). - PubMed
-
- Cayuela M. L., Elías-Arnanz M., Peñalver-Mellado M., Padmanabhan S. & Murillo F. J. The Stigmatella aurantiaca homolog of Myxococcus xanthus high-mobility-group A-type transcription factor CarD: insights into the functional modules of CarD and their distribution in bacteria. J. Bacteriol. 185, 3527–3537 (2003). - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

