Transcriptional mechanism of vascular endothelial growth factor-induced expression of protein kinase CβII in chronic lymphocytic leukaemia cells
- PMID: 28233872
- PMCID: PMC5324130
- DOI: 10.1038/srep43228
Transcriptional mechanism of vascular endothelial growth factor-induced expression of protein kinase CβII in chronic lymphocytic leukaemia cells
Abstract
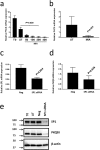
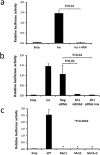
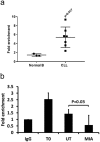
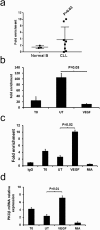
A key feature of chronic lymphocytic leukaemia (CLL) cells is overexpressed protein kinase CβII (PKCβII), an S/T kinase important in the pathogenesis of this and other B cell malignancies. The mechanisms contributing to enhanced transcription of the gene coding for PKCβII, PRKCB, in CLL cells remain poorly described, but could be important because of potential insight into how the phenotype of these cells is regulated. Here, we show that SP1 is the major driver of PKCβII expression in CLL cells where enhanced association of this transcription factor with the PRKCB promoter is likely because of the presence of histone marks permissive of gene activation. We also show how vascular endothelial growth factor (VEGF) regulates PRKCB promoter function in CLL cells, stimulating PKCβ gene transcription via increased association of SP1 and decreased association of STAT3. Taken together, these results are the first to demonstrate a clear role for SP1 in the up regulation of PKCβII expression in CLL cells, and the first to link SP1 with the pathogenesis of this and potentially other B cell malignancies where PKCβII is overexpressed.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
-
- Watson L., Wyld P. & Catovsky D. Disease burden of chronic lymphocytic leukaemia within the European Union. Eur J Haematol 81, 253–258 (2008). - PubMed
-
- Dighiero G. & Hamblin T. J. Chronic lymphocytic leukaemia. Lancet 371, 1017–1029 (2008). - PubMed
-
- Abrams S. T. et al.. B-cell receptor signaling in chronic lymphocytic leukemia cells is regulated by overexpressed active protein kinase CβII. Blood 109, 1193–1201 (2007). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

