Computational methods using genome-wide association studies to predict radiotherapy complications and to identify correlative molecular processes
- PMID: 28233873
- PMCID: PMC5324069
- DOI: 10.1038/srep43381
Computational methods using genome-wide association studies to predict radiotherapy complications and to identify correlative molecular processes
Abstract
The biological cause of clinically observed variability of normal tissue damage following radiotherapy is poorly understood. We hypothesized that machine/statistical learning methods using single nucleotide polymorphism (SNP)-based genome-wide association studies (GWAS) would identify groups of patients of differing complication risk, and furthermore could be used to identify key biological sources of variability. We developed a novel learning algorithm, called pre-conditioned random forest regression (PRFR), to construct polygenic risk models using hundreds of SNPs, thereby capturing genomic features that confer small differential risk. Predictive models were trained and validated on a cohort of 368 prostate cancer patients for two post-radiotherapy clinical endpoints: late rectal bleeding and erectile dysfunction. The proposed method results in better predictive performance compared with existing computational methods. Gene ontology enrichment analysis and protein-protein interaction network analysis are used to identify key biological processes and proteins that were plausible based on other published studies. In conclusion, we confirm that novel machine learning methods can produce large predictive models (hundreds of SNPs), yielding clinically useful risk stratification models, as well as identifying important underlying biological processes in the radiation damage and tissue repair process. The methods are generally applicable to GWAS data and are not specific to radiotherapy endpoints.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


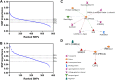
References
-
- Rattay T. & Talbot C. J. Finding the genetic determinants of adverse reactions to radiotherapy. Clin Oncol (R Coll Radiol) 26, 301–308 (2014). - PubMed
-
- Ringborg U. et al. The Swedish Council on Technology Assessment in Health Care (SBU) systematic overview of radiotherapy for cancer including a prospective survey of radiotherapy practice in Sweden 2001–summary and conclusions. Acta Oncol 42, 357–365 (2003). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

