Ubiquitin-like Protein Conjugation: Structures, Chemistry, and Mechanism
- PMID: 28234446
- PMCID: PMC5815371
- DOI: 10.1021/acs.chemrev.6b00737
Ubiquitin-like Protein Conjugation: Structures, Chemistry, and Mechanism
Abstract
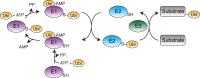
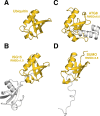
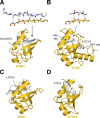
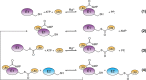
Ubiquitin-like proteins (Ubl's) are conjugated to target proteins or lipids to regulate their activity, stability, subcellular localization, or macromolecular interactions. Similar to ubiquitin, conjugation is achieved through a cascade of activities that are catalyzed by E1 activating enzymes, E2 conjugating enzymes, and E3 ligases. In this review, we will summarize structural and mechanistic details of enzymes and protein cofactors that participate in Ubl conjugation cascades. Precisely, we will focus on conjugation machinery in the SUMO, NEDD8, ATG8, ATG12, URM1, UFM1, FAT10, and ISG15 pathways while referring to the ubiquitin pathway to highlight common or contrasting themes. We will also review various strategies used to trap intermediates during Ubl activation and conjugation.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
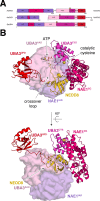
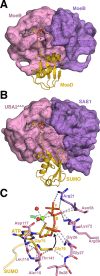
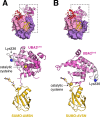
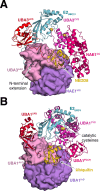

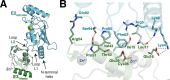
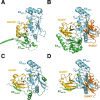
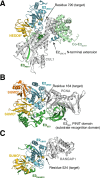
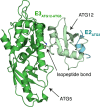
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

