One thousand somatic SNVs per skin fibroblast cell set baseline of mosaic mutational load with patterns that suggest proliferative origin
- PMID: 28235832
- PMCID: PMC5378170
- DOI: 10.1101/gr.215517.116
One thousand somatic SNVs per skin fibroblast cell set baseline of mosaic mutational load with patterns that suggest proliferative origin
Abstract
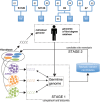
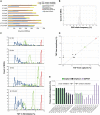
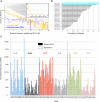
Few studies have been conducted to understand post-zygotic accumulation of mutations in cells of the healthy human body. We reprogrammed 32 skin fibroblast cells from families of donors into human induced pluripotent stem cell (hiPSC) lines. The clonal nature of hiPSC lines allows a high-resolution analysis of the genomes of the founder fibroblast cells without being confounded by the artifacts of single-cell whole-genome amplification. We estimate that on average a fibroblast cell in children has 1035 mostly benign mosaic SNVs. On average, 235 SNVs could be directly confirmed in the original fibroblast population by ultradeep sequencing, down to an allele frequency (AF) of 0.1%. More sensitive droplet digital PCR experiments confirmed more SNVs as mosaic with AF as low as 0.01%, suggesting that 1035 mosaic SNVs per fibroblast cell is the true average. Similar analyses in adults revealed no significant increase in the number of SNVs per cell, suggesting that a major fraction of mosaic SNVs in fibroblasts arises during development. Mosaic SNVs were distributed uniformly across the genome and were enriched in a mutational signature previously observed in cancers and in de novo variants and which, we hypothesize, is a hallmark of normal cell proliferation. Finally, AF distribution of mosaic SNVs had distinct narrow peaks, which could be a characteristic of clonal cell selection, clonal expansion, or both. These findings reveal a large degree of somatic mosaicism in healthy human tissues, link de novo and cancer mutations to somatic mosaicism, and couple somatic mosaicism with cell proliferation.
© 2017 Abyzov et al.; Published by Cold Spring Harbor Laboratory Press.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
