Adaptation of Candida albicans to Reactive Sulfur Species
- PMID: 28235888
- PMCID: PMC5419466
- DOI: 10.1534/genetics.116.199679
Adaptation of Candida albicans to Reactive Sulfur Species
Abstract
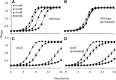
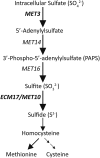
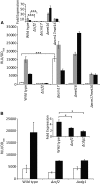
Candida albicans is an opportunistic fungal pathogen that is highly resistant to different oxidative stresses. How reactive sulfur species (RSS) such as sulfite regulate gene expression and the role of the transcription factor Zcf2 and the sulfite exporter Ssu1 in such responses are not known. Here, we show that C. albicans specifically adapts to sulfite stress and that Zcf2 is required for that response as well as induction of genes predicted to remove sulfite from cells and to increase the intracellular amount of a subset of nitrogen metabolites. Analysis of mutants in the sulfate assimilation pathway show that sulfite conversion to sulfide accounts for part of sulfite toxicity and that Zcf2-dependent expression of the SSU1 sulfite exporter is induced by both sulfite and sulfide. Mutations in the SSU1 promoter that selectively inhibit induction by the reactive nitrogen species (RNS) nitrite, a previously reported activator of SSU1, support a model for C. albicans in which Cta4-dependent RNS induction and Zcf2-dependent RSS induction are mediated by parallel pathways, different from S. cerevisiae in which the transcription factor Fzf1 mediates responses to both RNS and RSS. Lastly, we found that endogenous sulfite production leads to an increase in resistance to exogenously added sulfite. These results demonstrate that C. albicans has a unique response to sulfite that differs from the general oxidative stress response, and that adaptation to internal and external sulfite is largely mediated by one transcription factor and one effector gene.
Keywords: Candida albicans; ZCF2; sulfide; sulfite.
Copyright © 2017 by the Genetics Society of America.
Figures





References
-
- Avram D., Leid M., Bakalinsky A. T., 1999. Fzf1p of Saccharomyces cerevisiae is a positive regulator of SSU1 transcription and its first zinc finger region is required for DNA binding. Yeast 15: 473–480. - PubMed
-
- Bailey T. L., Elkan C., 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2: 28–36. - PubMed
-
- Bar-Even A., Flamholz A., Noor E., Milo R., 2012. Rethinking glycolysis: on the biochemical logic of metabolic pathways. Nat. Chem. Biol. 8: 509–517. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

