Two Acyltransferases Contribute Differently to Linolenic Acid Levels in Seed Oil
- PMID: 28235891
- PMCID: PMC5373062
- DOI: 10.1104/pp.16.01865
Two Acyltransferases Contribute Differently to Linolenic Acid Levels in Seed Oil
Abstract
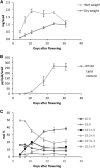
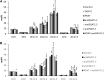
Acyltransferases are key contributors to triacylglycerol (TAG) synthesis and, thus, are of great importance for seed oil quality. The effects of increased or decreased expression of ACYL-COENZYME A:DIACYLGLYCEROL ACYLTRANSFERASE1 (DGAT1) or PHOSPHOLIPID:DIACYLGLYCEROL ACYLTRANSFERASE (PDAT) on seed lipid composition were assessed in several Camelina sativa lines. Furthermore, in vitro assays of acyltransferases in microsomal fractions prepared from developing seeds of some of these lines were performed. Decreased expression of DGAT1 led to an increased percentage of 18:3n-3 without any change in total lipid content of the seed. The tri-18:3 TAG increase occurred predominantly in the cotyledon, as determined with matrix-assisted laser desorption/ionization-mass spectrometry, whereas species with two 18:3n-3 acyl groups were elevated in both cotyledon and embryonal axis. PDAT overexpression led to a relative increase of 18:2n-6 at the expense of 18:3n-3, also without affecting the total lipid content. Differential distributions of TAG species also were observed in different parts of the seed. The microsomal assays revealed that C.sativa seeds have very high activity of diacylglycerol-phosphatidylcholine interconversion. The combination of analytical and biochemical data suggests that the higher 18:2n-6 content in the seed oil of the PDAT overexpressors is due to the channeling of fatty acids from phosphatidylcholine into TAG before being desaturated to 18:3n-3, caused by the high activity of PDAT in general and by PDAT specificity for 18:2n-6. The higher levels of 18:3n-3 in DGAT1-silencing lines are likely due to the compensatory activity of a TAG-synthesizing enzyme with specificity for this acyl group and more desaturation of acyl groups occurring on phosphatidylcholine.
© 2017 American Society of Plant Biologists. All Rights Reserved.
Figures







References
-
- Banas W, Carlsson AS, Banas A (2014) Effect of overexpression of PDAT gene on Arabidopsis growth rate and seed oil content. J Agric Sci 6: 65
-
- Bansal S, Durrett TP (2016) Camelina sativa: an ideal platform for the metabolic engineering and field production of industrial lipids. Biochimie 120: 9–16 - PubMed
-
- Bates PD, Browse J (2011) The pathway of triacylglycerol synthesis through phosphatidylcholine in Arabidopsis produces a bottleneck for the accumulation of unusual fatty acids in transgenic seeds. Plant J 68: 387–399 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

