Molecular assays for the diagnosis of sepsis in neonates
- PMID: 28236648
- PMCID: PMC6464551
- DOI: 10.1002/14651858.CD011926.pub2
Molecular assays for the diagnosis of sepsis in neonates
Update in
-
Molecular assays for the diagnosis of sepsis in neonates: a diagnostic test accuracy review.Cochrane Database Syst Rev. 2025 Mar 19;3(3):CD011926. doi: 10.1002/14651858.CD011926.pub3. Cochrane Database Syst Rev. 2025. PMID: 40105375
Abstract
Background: Microbial cultures for diagnosis of neonatal sepsis have low sensitivity and reporting delay. Advances in molecular microbiology have fostered new molecular assays that are rapid and may improve neonatal outcomes.
Objectives: To assess the diagnostic accuracy of various molecular methods for the diagnosis of culture-positive bacterial and fungal sepsis in neonates and to explore heterogeneity among studies by analyzing subgroups classified by gestational age and type of sepsis onset and compare molecular tests with one another.
Search methods: We performed the systematic review as recommended by the Cochrane Diagnostic Test Accuracy Working Group. On 19 January 2016, we searched electronic bibliographic databases (the Cochrane Library, PubMed (from 1966), Embase (from 1982), and CINAHL (from 1982)), conference proceedings of the Pediatric Academic Societies annual conference (from 1990), clinical trial registries (ClinicalTrials.gov, International Standard Randomised Controlled Trial Number (ISRCTN) registry, and World Health Organization (WHO) International Clinical Trials Platform (ICTRP) Search portal), and Science Citation Index. We contacted experts in the field for studies.
Selection criteria: We included studies that were prospective or retrospective, cohort or cross-sectional design, which evaluated molecular assays (index test) in neonates with suspected sepsis (participants) in comparison with microbial cultures (reference standard).
Data collection and analysis: Two review authors independently assessed the methodologic quality of the studies and extracted data. We performed meta-analyses using the bivariate and hierarchical summary receiver operating characteristic (HSROC) models and entered data into Review Manager 5.
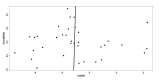
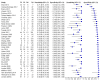
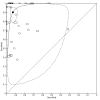
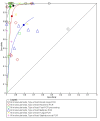
Main results: Thirty-five studies were eligible for inclusion and the summary estimate of sensitivity was 0.90 (95% confidence interval (CI) 0.82 to 0.95) and of specificity was 0.93 (95% CI 0.89 to 0.96) (moderate quality evidence). We explored heterogeneity by subgroup analyses of type of test, gestational age, type of sepsis onset, and prevalence of sepsis and we did not find sufficient explanations for the heterogeneity (moderate to very low quality evidence). Sensitivity analyses by including studies that analyzed blood samples and by good methodology revealed similar results (moderate quality evidence).
Authors' conclusions: Molecular assays have the advantage of producing rapid results and may perform well as 'add-on' tests.
Conflict of interest statement
Mohan Pammi, Angela Flores, James Versalovic and Mariska MG Leeflang have no financial or other conflicts of interest to disclose.
Figures




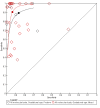



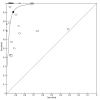

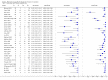

References
References to studies included in this review
Briones 2003 {published data only}
-
- Briones CR, Villanueva‐Uy ME, Uy HGTI. The use of polymerase chain reaction in neonatal candidemia. Pediatric Research 2003;53:396A.
Chan 2009 {published data only}
-
- Chan KY, Lam HS, Cheung HM, Chan AK, Li K, Fok TF, et al. Rapid identification and differentiation of Gram‐negative and Gram‐positive bacterial bloodstream infections by quantitative polymerase chain reaction in preterm infants. Critical Care Medicine 2009;37:2441‐7. [PUBMED: 19531943] - PubMed
Chen 2009 {published data only}
-
- Chen LH, Duan QJ, Cai MT, Wu YD, Shang SQ. Rapid diagnosis of sepsis and bacterial meningitis in children with real‐time fluorescent quantitative polymerase chain reaction amplification in the bacterial 16S rRNA gene. Clinical Pediatrics 2009;48(6):641‐7. [PUBMED: 19407210] - PubMed
Draz 2013 {published data only}
-
- Draz NI, Taha SE, Abou Shady NM, Abdel Ghany YS. Comparison of broad range 16S rDNA PCR to conventional blood culture for diagnosis of sepsis in the newborn. Egyptian Journal of Human Medical Genetics 2013;14:403‐11. [PUBMED: 19152691]
Dutta 2009 {published data only}
-
- Dutta S, Narang A, Chakraborty A, Ray P. Diagnosis of neonatal sepsis using universal primer polymerase chain reaction before and after starting antibiotic drug therapy. Archives of Pediatrics and Adolescent Medicine 2009;163(1):6‐11. [PUBMED: 19124696] - PubMed
Enomoto 2009 {published data only}
-
- Enomoto M, Morioka I, Morisawa T, Yokoyama N, Matsuo M. A novel diagnostic tool for detecting neonatal infections using multiplex polymerase chain reaction. Neonatology 2009;96(2):102‐8. [PUBMED: 19279393] - PubMed
Esparcia 2011 {published data only}
-
- Esparcia O, Montemayor M, Ginovart G, Pomar V, Soriano G, Pericas R, et al. Diagnostic accuracy of a 16S ribosomal DNA gene‐based molecular technique (RT‐PCR, microarray, and sequencing) for bacterial meningitis, early‐onset neonatal sepsis, and spontaneous bacterial peritonitis. Diagnostic Microbiology and Infectious Disease 2011;69(2):153‐60. [PUBMED: 21251558] - PubMed
Fujimori 2010 {published data only}
Garcia‐Elorriaga 2012 {published data only}
-
- Garcia‐Elorriaga G, Cortes‐Torres N, Ballesteros‐Del‐Olmo JC, Rey‐Pineda G, Gonzaez‐Bonilla C. The usefulness of the buffy coat smear and panbacterial polymerase chain reaction in early diagnosis of neonatal sepsis. Revista de Investigacion Clinica 2012;64(3):275‐83. [PUBMED: 23045950] - PubMed
Ibarra 2015 {published data only}
Jordan 2000 {published data only}
Jordan 2005a {published data only}
Jordan 2006 {published data only}
Kasper 2013 {published data only}
-
- Kasper DC, Altiok I, Mechtler TP, Bohm J, Straub J, Langgartner M, et al. Molecular detection of late‐onset neonatal sepsis in premature infants using small blood volumes: proof‐of‐concept. Neonatology 2013;103(4):268‐73. [PUBMED: 23485823] - PubMed
Laforgia 1997 {published data only}
-
- Laforgia N, Coppola B, Carbone R, Grassi A, Mautone A, Lolascon A. Rapid detection of neonatal sepsis using polymerase chain reaction. Acta Paediatrica 1997;86(10):1097‐9. [PUBMED: 9350892] - PubMed
Lima 2007 {published data only}
-
- Lima V, Alpuche A, Noyola D, Soria R, Nieto K. Polymerase chain reaction technique in the diagnosis of neonatal sepsis: future gold standard?. Pediatric Academic Societies Annual Meeting; 2007 May 5‐8; Toronto (ON) 2007.
Liu 2014 {published data only}
-
- Liu CL, Ai HW, Wang WP, Chen L, Hu HB, Ye T, et al. Comparison of 16S rRNA gene PCR and blood culture for diagnosis of neonatal sepsis. Archives of Pediatrics 2014;21(2):162‐9. [PUBMED: 24388336] - PubMed
Makhoul 2005 {published data only}
Makhoul 2006 {published data only}
-
- Makhoul IR, Yacoub A, Smolkin T, Sujov P, Kassis I, Sprecher H. Values of C‐reactive protein, procalcitonin, and Staphylococcus‐specific PCR in neonatal late‐onset sepsis. Acta Paediatrica 2006;95(10):1218‐23. [PUBMED: 16982493] - PubMed
Ohlin 2008 {published data only}
-
- Ohlin A, Backman A, Bjorkqvist M, Molling P, Jurstrand M, Schollin J. Real‐time PCR of the 16S‐rRNA gene in the diagnosis of neonatal bacteraemia. Acta Paediatrica 2008;97(10):1376‐80. [PUBMED: 18624992] - PubMed
Ohlin 2012 {published data only}
-
- Ohlin A, Backman A, Ewald U, Schollin J, Bjorkqvist M. Diagnosis of neonatal sepsis by broad‐range 16S real‐time polymerase chain reaction. Neonatology 2012;10(4):241‐6. [PUBMED: 22205207] - PubMed
Paolucci 2009 {published data only}
-
- Paolucci M, Capretti MG, Dal Monte P, Corvaglia L, Landini MP, Varani S, et al. Laboratory diagnosis of late‐onset sepsis in newborns by multiplex real‐time PCR. Journal of Medical Microbiology 2009;58(Pt 4):533‐4. [PUBMED: 19273654] - PubMed
Reier‐Nilsen 2009 {published data only}
Shaat 2013 {published data only}
-
- Shaat SS, Shazly SA, Badr Eldin MM, Barakat SS, Hashish MH. Role of polymerase chain reaction as an early diagnostic tool for neonatal bacterial sepsis. Journal of Egypt Public Health Association 2013;88(3):160‐4. [PUBMED: 24374951] - PubMed
Shang 2005 {published data only}
-
- Shang S, Chen G, Wu Y, Du L, Zhao Z. Rapid diagnosis of bacterial sepsis with PCR amplification and microarray hybridization in 16S rRNA gene. Pediatric Research 2005;58(1):143‐8. [PUBMED: 15985688] - PubMed
Taira 2014 {published data only}
Tirodker 2003 {published data only}
-
- Tirodker UH, Nataro JP, Smith S, LasCasas L, Fairchild KD. Detection of fungemia by polymerase chain reaction in critically ill neonates and children. Journal of Perinatology 2003;23(2):117‐22. [PUBMED: 12673260] - PubMed
Tong 2004 {published data only}
-
- Tong MQ, Shang SQ, Wu YD, Zhao ZY. Rapid diagnosis of neonatal sepsis by 16SrRNA genes PCR amplification and genechip hybridization. Zhonghua Er Ke Za Zhi 2004;42(9):663‐7. [PUBMED: 15482666] - PubMed
Torres‐Martos 2013 {published data only}
-
- Torres‐Martos E, Perez‐Ruiz M, Pedrosa‐Corral I, Pena‐Caballero M, Jimenez‐Valera MM, Perez‐Ramirez MD, et al. Evaluation of the LightCycler® SeptiFast test in newborns and infants with clinical suspicion of sepsis. Enfermedades Infecciosas Microbiologia Clinica 2013;31(6):375‐9. [PUBMED: 23137657] - PubMed
Trovato 2012 {published data only}
-
- Trovato L, Betta P, Romeo MG, Oliveri S. Detection of fungal DNA in lysis‐centrifugation blood culture for the diagnosis of invasive candidiasis in neonatal patients. Clinical Microbiology and Infection 2012;18(3):E63‐5. [PUBMED: 22192484] - PubMed
Van der Brand 2014 {published data only}
-
- Brand M, Peters RP, Catsburg A, Rubenjan A, Broeke FJ, Dungen FA, et al. Development of a multiplex real‐time PCR assay for the rapid diagnosis of neonatal late onset sepsis. Journal of Microbiological Methods 2014;106:8‐15. [PUBMED: 25102109] - PubMed
Villanueva‐Uy 2003 {published data only}
-
- Villanueva‐Uy ME, Briones CR, Uy HG. Application of polymerase chain reaction in late‐onset neonatal sepsis. Pediatric Research 2003;53:313A.
Wu 2007 {published data only}
-
- Wu YD, Shang SQ, Li JP, Yang ZQ, Zheng ZB, Du LZ, et al. A broad‐range 16S rRNA gene real‐time PCR assay for the diagnosis of neonatal septicemia. Zhonghua Er Ke Za Zhi 2007;45(6):446‐9. [PUBMED: 17880793] - PubMed
Wu 2008 {published data only}
Yadav 2005 {published data only}
-
- Yadav AK, Wilson CG, Prasad PL, Menon PK. Polymerase chain reaction in rapid diagnosis of neonatal sepsis. Indian Pediatrics 2005;42(7):681‐5. [PUBMED: 16085969] - PubMed
References to studies excluded from this review
Chiba 2009 {published data only}
-
- Chiba N, Murayama SY, Morozumi M, Nakayama E, Okada T, Iwata S, et al. Rapid detection of eight causative pathogens for the diagnosis of bacterial meningitis by real‐time PCR. Journal of Infection and Chemotherapy 2009;15(2):92‐8. [PUBMED: 19396518] - PubMed
Das 2015 {published data only}
-
- Das BK, Suri S, Nath G, Prasad R. Urine nested polymerase chain reaction in neonatal septicemia. Journal of Tropical Pediatrics 2015;61(4):295‐300. [PUBMED: 26130622] - PubMed
de Zoysa 2012 {published data only}
-
- Zoysa A, Edwards K, Gharbia S, Underwood A, Charlett A, Efstratiou A. Non‐culture detection of Streptococcus agalactiae (Lancefield group B Streptococcus) in clinical samples by real‐time PCR. Journal of Medical Microbiology 2012;61(Pt 8):1086‐90. [PUBMED: 22740612] - PubMed
Golden 2004 {published data only}
-
- Golden SM, Stamilio DM, Faux BM, dela Cruz WP, Shoemaker CT, Blackmon CL, et al. Evaluation of a real‐time fluorescent PCR assay for rapid detection of Group B Streptococci in neonatal blood. Diagnostic Microbiology and Infectious Disease 2004;50(1):7‐13. [PUBMED: 15380273] - PubMed
Jones 2010 {published data only}
-
- Jones V, Wilks M, Johnson G, Warwick S, Hennessey E, Kempley S, et al. The use of molecular techniques for bacterial detection in the analysis of gastric aspirates collected from infants on the first day of life. Early Human Development 2010;86(3):167‐70. [PUBMED: 20223606] - PubMed
Jordan 2005b {published data only}
Jordan 2009 {published data only}
Lucignano 2011 {published data only}
Makhoul 2007 {published data only}
-
- Makhoul IR, Sprecher H, Smolkin T, Sawaid R, Ben‐David S, Sujov P, et al. Approach to term neonates born after maternal intrapartum fever and unknown maternal group B Streptococcus status: value of serum C‐reactive protein and 16S rRNA gene PCR amplification. Pediatric Infectious Disease Journal 2007;26(11):1064‐6. [PUBMED: 17984819] - PubMed
Shang 2001 {published data only}
-
- Shang S, Chen Z, Yu X. Detection of bacterial DNA by PCR and reverse hybridization in the 16S rRNA gene with particular reference to neonatal septicemia. Acta Paediatrica 2001;90(2):179‐83. [PUBMED: 11236048] - PubMed
Shen 2004 {published data only}
-
- Shen DX, Du J, Feng ZC. Rapid diagnosis of common pathogenic bacteria infection in newborn infants by 16SrDNA oligonucleotide array. Zhonghua Er Ke Za Zhi 2004;42(9):668‐72. [PUBMED: 15482667] - PubMed
Tschiedel 2012 {published data only}
-
- Tschiedel E, Steinmann J, Buer J, Onnebrink JG, Felderhoff‐Muser U, Rath PM, et al. Results and relevance of molecular detection of pathogens by SeptiFast ‐ a retrospective analysis in 75 critically ill children. Klinische Padiatrie 2012;224(1):12‐6. [PUBMED: 22258624] - PubMed
Additional references
Adams‐Chapman 2006
-
- Adams‐Chapman I, Stoll BJ. Neonatal infection and long‐term neurodevelopmental outcome in the preterm infant. Current Opinion in Infectious Diseases 2006;19(3):290‐7. [PUBMED: 16645492] - PubMed
Blommendahl 2002
-
- Blommendahl J, Janas M, Laine S, Miettinen A, Ashorn P. Comparison of procalcitonin with CRP and differential white blood cell count for diagnosis of culture‐proven neonatal sepsis. Scandinavian Journal of Infectious Diseases 2002;34(8):620‐2. [PUBMED: 12238581] - PubMed
Bossuyt 2006
Brozanski 2006
-
- Brozanski BS, Jones JG, Krohn MJ, Jordan JA. Use of polymerase chain reaction as a diagnostic tool for neonatal sepsis can result in a decrease in use of antibiotics and total neonatal intensive care unit length of stay. Journal of Perinatology 2006;26(11):688‐692. - PubMed
Canadian Neonatal Network 2014
-
- Shah P, Yoon EW, Chan P, Members of the Annual Report Review Committee. Annual report of the Canadian Neonatal Network, 2004. www.canadianneonatalnetwork.org/Portal/LinkClick.aspx?fileticket=eGgxmMu... (accessed 18 February 2017).
Chiesa 2004
-
- Chiesa C, Panero A, Osborn JF, Simonetti AF, Pacifico L. Diagnosis of neonatal sepsis: a clinical and laboratory challenge. Clinical Chemistry 2004;50(2):279‐87. [PUBMED: 14752012] - PubMed
Dong 2015
Gopalakrishna 2014
-
- Gopalakrishna G, Mustafa RA, Davenport C, Scholten RJ, Hyde C, Brozek J, et al. Applying Grading of Recommendations Assessment, Development and Evaluation (GRADE) to diagnostic tests was challenging but doable. Journal of Clinical Epidemiology 2014;67(7):760‐8. - PubMed
Harbord 2007
-
- Harbord RM, Deeks JJ, Egger M, Whiting P, Sterne JA. A unification of models for meta‐analysis of diagnostic accuracy studies. Biostatistics 2007;8(2):239‐51. [PUBMED: 16698768] - PubMed
Harbord 2008
-
- Harbord RM, Whiting P, Sterne JA, Egger M, Deeks JJ, Shang A, et al. An empirical comparison of methods for meta‐analysis of diagnostic accuracy showed hierarchical models are necessary. Journal of Clinical Epidemiology 2008;61(11):1095‐103. [PUBMED: 19208372] - PubMed
Harbord 2009
-
- Harbord RM, Whiting P. Metandi: meta‐analysis of diagnostic accuracy using hierarchical logistic regression. Stata Journal 2009;9(2):211‐29.
Hornik 2012
Isaacman 1996
-
- Isaacman DJ, Karasic RB, Reynolds EA, Kost SI. Effect of number of blood cultures and volume of blood on detection of bacteremia in children. Pediatrics 1996;128(2):190‐5. [PUBMED: 8636810] - PubMed
Jordan 2010
-
- Jordan JA. Molecular diagnosis of neonatal sepsis. Clinics in Perinatology 2010;37(2):411‐9. [PUBMED: 20569815] - PubMed
Leeflang 2008
Macaskill 2010
-
- Macaskill P, Gatsonis C, Deeks JJ, Harbord RM, Takwoingi Y. Chapter 10: Analysing and presenting results. In: Cochrane Handbook for Systematic Reviews of Diagnostic Test Accuracy Version 1.0.0. The Cochrane Collaboration, 2010. Available from srdta.cochrane.org/.
Ng 1997
-
- Ng PC, Cheng SH, Chui KM, Fok TF, Wong MY, Wong W, et al. Diagnosis of late onset neonatal sepsis with cytokines, adhesion molecule, and C‐reactive protein in preterm very low birthweight infants. Archives of Disease in Childhood Fetal and Neonatal Edition 1997;77(3):F221‐7. [PUBMED: 9462194] - PMC - PubMed
Ng 2012
-
- Ng PC, Lam HS. Biomarkers in neonatology: the next generation of tests. Neonatology 2012;102(2):145‐51. [PUBMED: 22759988] - PubMed
Pammi 2011
-
- Pammi M, Flores A, Leeflang M, Versalovic J. Molecular assays in the diagnosis of neonatal sepsis: a systematic review and meta‐analysis. Pediatrics 2011;128(4):e973‐85. [PUBMED: 21949139] - PubMed
Reitsma 2009
-
- Reitsma JB, Whiting P, Vlassov VV, Leeflang MMG, Deeks JJ. Chapter 9: Assessing methodological quality. In: Cochrane Handbook for Systematic Reviews of Diagnostic Test Accuracy Version 1.0.0. The Cochrane Collaboration, 2009. Available from srdta.cochrane.org/.
Relman 1999
-
- Relman DA. The search for unrecognized pathogens. Science 1999;284(5418):1308‐10. [PUBMED: 10334977] - PubMed
RevMan 2014 [Computer program]
-
- The Nordic Cochrane Centre, The Cochrane Collaboration. Review Manager (RevMan). Version 5.3. Copenhagen: The Nordic Cochrane Centre, The Cochrane Collaboration, 2014.
Rodriguez‐Creixems 2008
-
- Rodriguez‐Creixems M, Alcala L, Munoz P, Cercenado E, Vicente T, Bouza E. Bloodstream infections: evolution and trends in the microbiology workload, incidence, and etiology, 1985‐2006. Medicine 2008;87(4):234‐49. [PUBMED: 18626306] - PubMed
Schelonka 1996
-
- Schelonka RL, Chai MK, Yoder BA, Hensley D, Brockett RM, Ascher DP. Volume of blood required to detect common neonatal pathogens. Journal of Pediatrics 1996;129(2):275‐8. [PUBMED: 8765627] - PubMed
Schünemann 2016
-
- Schünemann HJ, Mustafa R, Brozek J, Santesso N, Alonso‐Coello P, Guyatt G, et al. GRADE Working Group. GRADE Guidelines: 16. GRADE evidence to decision frameworks for tests in clinical practice and public health. Journal of Clinical Epidemiology 2016;76:89‐98. - PubMed
Stata 2011 [Computer program]
-
- StataCorp. LP. Stata Statistical Software: Release 11. College Station (TX): StataCorp. LP, 2011.
Stoll 2002
-
- Stoll BJ, Hansen N, Fanaroff AA, Wright LL, Carlo WA, Ehrenkranz RA, et al. Late‐onset sepsis in very low birth weight neonates: the experience of the NICHD Neonatal Research Network. Pediatrics 2002;110(2 Pt 1):285‐91. [PUBMED: 12165580] - PubMed
Stoll 2004
-
- Stoll BJ, Hansen NI, Adams‐Chapman I, Fanaroff AA, Hintz SR, Vohr B, et al. Neurodevelopmental and growth impairment among extremely low‐birth‐weight infants with neonatal infection. JAMA 2004;292(19):2357‐65. [PUBMED: 15547163] - PubMed
Trenti 2016
-
- Trenti T, Schunemann HJ, Plebani M. Developing GRADE outcome‐based recommendations about diagnostic tests: a key role in laboratory medicine policies. Clinical Chemistry and Laboratory Medicine 2016;54(4):535‐43. - PubMed
Van Enst 2014
Venkatesh 2010
Verboon‐Maciolek 2006
-
- Verboon‐Maciolek MA, Thijsen SF, Hemels MA, Menses M, Loon AM, Krediet TG, et al. Inflammatory mediators for the diagnosis and treatment of sepsis in early infancy. Pediatric Research 2006;59(3):457‐61. [PUBMED: 16492989] - PubMed
Whiting 2011
-
- Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS‐2: a revised tool for the quality assessment of diagnostic accuracy studies. Annals of Internal Medicine 2011;155(8):529‐36. [PUBMED: 22007046] - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

