Permanganate/S1 Nuclease Footprinting Reveals Non-B DNA Structures with Regulatory Potential across a Mammalian Genome
- PMID: 28237796
- PMCID: PMC7432990
- DOI: 10.1016/j.cels.2017.01.013
Permanganate/S1 Nuclease Footprinting Reveals Non-B DNA Structures with Regulatory Potential across a Mammalian Genome
Abstract
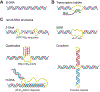
DNA in cells is predominantly B-form double helix. Though certain DNA sequences in vitro may fold into other structures, such as triplex, left-handed Z form, or quadruplex DNA, the stability and prevalence of these structures in vivo are not known. Here, using computational analysis of sequence motifs, RNA polymerase II binding data, and genome-wide potassium permanganate-dependent nuclease footprinting data, we map thousands of putative non-B DNA sites at high resolution in mouse B cells. Computational analysis associates these non-B DNAs with particular structures and indicates that they form at locations compatible with an involvement in gene regulation. Further analyses support the notion that non-B DNA structure formation influences the occupancy and positioning of nucleosomes in chromatin. These results suggest that non-B DNAs contribute to the control of a variety of critical cellular and organismal processes.
Keywords: DNA topology; H-DNA; SIDD; Z-DNA; chromatin; cruciform; non-B DNA; quadruplex; supercoiling; transcription.
Published by Elsevier Inc.
Figures

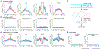



References
-
- Bacolla A, and Wells RD (2004). Non-B DNA conformations, genomic rearrangements, and human disease. The Journal of biological chemistry 279, 47411–47414. - PubMed
-
- Belotserkovskii BP, De Silva E, Tornaletti S, Wang G, Vasquez KM, and Hanawalt PC (2007). A triplex-forming sequence from the human c-MYC promoter interferes with DNA transcription. The Journal of biological chemistry 282, 32433–32441. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

