Channel of viral DNA packaging motor for real time kinetic analysis of peptide oxidation states
- PMID: 28237908
- PMCID: PMC5421631
- DOI: 10.1016/j.biomaterials.2017.01.031
Channel of viral DNA packaging motor for real time kinetic analysis of peptide oxidation states
Abstract
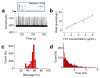
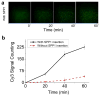
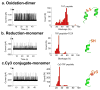
Nanopore technology has become a powerful tool in single molecule sensing, and protein nanopores appear to be more advantageous than synthetic counterparts with regards to channel amenability, structure homogeneity, and production reproducibility. However, the diameter of most of the well-studied protein nanopores is too small to allow the passage of protein or peptides that are typically in multiple nanometers scale. The portal channel from bacteriophage SPP1 has a large channel size that allows the translocation of peptides with higher ordered structures. Utilizing single channel conductance assay and optical single molecule imaging, we observed translocation of peptides and quantitatively analyzed the dynamics of peptide oligomeric states in real-time at single molecule level. The oxidative and the reduced states of peptides were clearly differentiated based on their characteristic electronic signatures. A similar Gibbs free energy (ΔG0) was obtained when different concentrations of substrates were applied, suggesting that the use of SPP1 nanopore for real-time quantification of peptide oligomeric states is feasible. With the intrinsic nature of size and conjugation amenability, the SPP1 nanopore has the potential for development into a tool for the quantification of peptide and protein structures in real time.
Keywords: Bacteriophage assembly; Biomotor; Nanobiotechnology; Nanopore sensing; Peptide identification; Viral motor.
Copyright © 2017 Elsevier Ltd. All rights reserved.
Figures

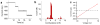

Similar articles
-
Fingerprinting of Peptides with a Large Channel of Bacteriophage Phi29 DNA Packaging Motor.Small. 2016 Sep;12(33):4572-8. doi: 10.1002/smll.201601157. Epub 2016 Jul 20. Small. 2016. PMID: 27435806 Free PMC article.
-
Oriented single directional insertion of nanochannel of bacteriophage SPP1 DNA packaging motor into lipid bilayer via polar hydrophobicity.Biomaterials. 2016 Oct;105:222-227. doi: 10.1016/j.biomaterials.2016.08.002. Epub 2016 Aug 4. Biomaterials. 2016. PMID: 27529454 Free PMC article.
-
Nano-channel of viral DNA packaging motor as single pore to differentiate peptides with single amino acid difference.Biomaterials. 2018 Nov;182:227-233. doi: 10.1016/j.biomaterials.2018.08.005. Epub 2018 Aug 3. Biomaterials. 2018. PMID: 30138785 Free PMC article.
-
Structure, assembly, and DNA packaging of the bacteriophage T4 head.Adv Virus Res. 2012;82:119-53. doi: 10.1016/B978-0-12-394621-8.00018-2. Adv Virus Res. 2012. PMID: 22420853 Free PMC article. Review.
-
Single-molecule studies of viral DNA packaging.Curr Opin Virol. 2011 Aug;1(2):134-41. doi: 10.1016/j.coviro.2011.05.023. Epub 2011 Jul 1. Curr Opin Virol. 2011. PMID: 22440623 Free PMC article. Review.
Cited by
-
Biomotors, viral assembly, and RNA nanobiotechnology: Current achievements and future directions.Comput Struct Biotechnol J. 2022 Nov 11;20:6120-6137. doi: 10.1016/j.csbj.2022.11.007. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 36420155 Free PMC article. Review.
-
DNA/RNA hybrids avoid channel gating that leads to the continued packaging of numerous hybrids into the phi29 protein shell.Nucleic Acids Res. 2025 Mar 20;53(6):gkaf242. doi: 10.1093/nar/gkaf242. Nucleic Acids Res. 2025. PMID: 40193705 Free PMC article.
-
Porphyrin-Assisted Docking of a Thermophage Portal Protein into Lipid Bilayers: Nanopore Engineering and Characterization.ACS Nano. 2017 Dec 26;11(12):11931-11945. doi: 10.1021/acsnano.7b06980. Epub 2017 Nov 15. ACS Nano. 2017. PMID: 29120602 Free PMC article.
-
Systemic Overview of Microstrip Patch Antenna's for Different Biomedical Applications.Adv Pharm Bull. 2021 May;11(3):439-449. doi: 10.34172/apb.2021.051. Epub 2020 Jul 1. Adv Pharm Bull. 2021. PMID: 34513618 Free PMC article. Review.
-
Voltage controlled shutter regulates channel size and motion direction of protein aperture as durable nano-electric rectifier-----An opinion in biomimetic nanoaperture.Biomaterials. 2022 Dec;291:121863. doi: 10.1016/j.biomaterials.2022.121863. Epub 2022 Nov 2. Biomaterials. 2022. PMID: 36356474 Free PMC article.
References
-
- Cuervo A, Carrascosa JL. Viral connectors for DNA encapsulation. Curr Opin Biotechnol. 2011;23:529–36. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

