PRMT5-Dependent Methylation of the TIP60 Coactivator RUVBL1 Is a Key Regulator of Homologous Recombination
- PMID: 28238654
- PMCID: PMC5344794
- DOI: 10.1016/j.molcel.2017.01.019
PRMT5-Dependent Methylation of the TIP60 Coactivator RUVBL1 Is a Key Regulator of Homologous Recombination
Abstract
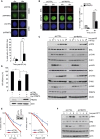
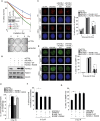
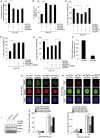
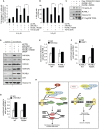
Protein post-translation modification plays an important role in regulating DNA repair; however, the role of arginine methylation in this process is poorly understood. Here we identify the arginine methyltransferase PRMT5 as a key regulator of homologous recombination (HR)-mediated double-strand break (DSB) repair, which is mediated through its ability to methylate RUVBL1, a cofactor of the TIP60 complex. We show that PRMT5 targets RUVBL1 for methylation at position R205, which facilitates TIP60-dependent mobilization of 53BP1 from DNA breaks, promoting HR. Mechanistically, we demonstrate that PRMT5-directed methylation of RUVBL1 is critically required for the acetyltransferase activity of TIP60, promoting histone H4K16 acetylation, which facilities 53BP1 displacement from DSBs. Interestingly, RUVBL1 methylation did not affect the ability of TIP60 to facilitate ATM activation. Taken together, our findings reveal the importance of PRMT5-mediated arginine methylation during DSB repair pathway choice through its ability to regulate acetylation-dependent control of 53BP1 localization.
Keywords: 52BP1; DNA repair; PRMT5; RUVBL1; TIP60; arginine methylation; homologous recombination.
Copyright © 2017 The Authors. Published by Elsevier Inc. All rights reserved.
Figures




Similar articles
-
In silico protein structural analysis of PRMT5 and RUVBL1 mutations arising in human cancers.Cancer Genet. 2025 Apr;292-293:49-56. doi: 10.1016/j.cancergen.2025.01.002. Epub 2025 Jan 17. Cancer Genet. 2025. PMID: 39874873 Free PMC article.
-
The TIP60 Complex Regulates Bivalent Chromatin Recognition by 53BP1 through Direct H4K20me Binding and H2AK15 Acetylation.Mol Cell. 2016 May 5;62(3):409-421. doi: 10.1016/j.molcel.2016.03.031. Mol Cell. 2016. PMID: 27153538 Free PMC article.
-
DNA double-strand breaks promote methylation of histone H3 on lysine 9 and transient formation of repressive chromatin.Proc Natl Acad Sci U S A. 2014 Jun 24;111(25):9169-74. doi: 10.1073/pnas.1403565111. Epub 2014 Jun 9. Proc Natl Acad Sci U S A. 2014. PMID: 24927542 Free PMC article.
-
Role of histone acetyltransferases MOF and Tip60 in genome stability.DNA Repair (Amst). 2021 Nov;107:103205. doi: 10.1016/j.dnarep.2021.103205. Epub 2021 Aug 8. DNA Repair (Amst). 2021. PMID: 34399315 Review.
-
Tip60: connecting chromatin to DNA damage signaling.Cell Cycle. 2010 Mar 1;9(5):930-6. doi: 10.4161/cc.9.5.10931. Epub 2010 Mar 11. Cell Cycle. 2010. PMID: 20160506 Free PMC article. Review.
Cited by
-
How Protein Methylation Regulates Steroid Receptor Function.Endocr Rev. 2022 Jan 12;43(1):160-197. doi: 10.1210/endrev/bnab014. Endocr Rev. 2022. PMID: 33955470 Free PMC article. Review.
-
PRMT5 promotes DNA repair through methylation of 53BP1 and is regulated by Src-mediated phosphorylation.Commun Biol. 2020 Aug 5;3(1):428. doi: 10.1038/s42003-020-01157-z. Commun Biol. 2020. PMID: 32759981 Free PMC article.
-
Protein arginine methylation: from enigmatic functions to therapeutic targeting.Nat Rev Drug Discov. 2021 Jul;20(7):509-530. doi: 10.1038/s41573-021-00159-8. Epub 2021 Mar 19. Nat Rev Drug Discov. 2021. PMID: 33742187 Review.
-
PRMT5 control of cGAS/STING and NLRC5 pathways defines melanoma response to antitumor immunity.Sci Transl Med. 2020 Jul 8;12(551):eaaz5683. doi: 10.1126/scitranslmed.aaz5683. Sci Transl Med. 2020. PMID: 32641491 Free PMC article.
-
Genome-wide R-loop analysis defines unique roles for DDX5, XRN2, and PRMT5 in DNA/RNA hybrid resolution.Life Sci Alliance. 2020 Aug 3;3(10):e202000762. doi: 10.26508/lsa.202000762. Print 2020 Oct. Life Sci Alliance. 2020. PMID: 32747416 Free PMC article.
References
-
- Arnaudeau C., Lundin C., Helleday T. DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism in mammalian cells. J. Mol. Biol. 2001;307:1235–1245. - PubMed
-
- Auclair Y., Richard S. The role of arginine methylation in the DNA damage response. DNA Repair (Amst.) 2013;12:459–465. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

