Mimivirus: leading the way in the discovery of giant viruses of amoebae
- PMID: 28239153
- PMCID: PMC7096837
- DOI: 10.1038/nrmicro.2016.197
Mimivirus: leading the way in the discovery of giant viruses of amoebae
Abstract
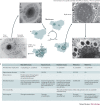
The accidental discovery of the giant virus of amoeba - Acanthamoeba polyphaga mimivirus (APMV; more commonly known as mimivirus) - in 2003 changed the field of virology. Viruses were previously defined by their submicroscopic size, which probably prevented the search for giant viruses, which are visible by light microscopy. Extended studies of giant viruses of amoebae revealed that they have genetic, proteomic and structural complexities that were not thought to exist among viruses and that are comparable to those of bacteria, archaea and small eukaryotes. The giant virus particles contain mRNA and more than 100 proteins, they have gene repertoires that are broader than those of other viruses and, notably, some encode translation components. The infection cycles of giant viruses of amoebae involve virus entry by amoebal phagocytosis and replication in viral factories. In addition, mimiviruses are infected by virophages, defend against them through the mimivirus virophage resistance element (MIMIVIRE) system and have a unique mobilome. Overall, giant viruses of amoebae, including mimiviruses, marseilleviruses, pandoraviruses, pithoviruses, faustoviruses and molliviruses, challenge the definition and classification of viruses, and have increasingly been detected in humans.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

