Biodegradation of alkaline lignin by Bacillus ligniniphilus L1
- PMID: 28239416
- PMCID: PMC5320714
- DOI: 10.1186/s13068-017-0735-y
Biodegradation of alkaline lignin by Bacillus ligniniphilus L1
Abstract
Background: Lignin is the most abundant aromatic biopolymer in the biosphere and it comprises up to 30% of plant biomass. Although lignin is the most recalcitrant component of the plant cell wall, still there are microorganisms able to decompose it or degrade it. Fungi are recognized as the most widely used microbes for lignin degradation. However, bacteria have also been known to be able to utilize lignin as a carbon or energy source. Bacillus ligniniphilus L1 was selected in this study due to its capability to utilize alkaline lignin as a single carbon or energy source and its excellent ability to survive in extreme environments.
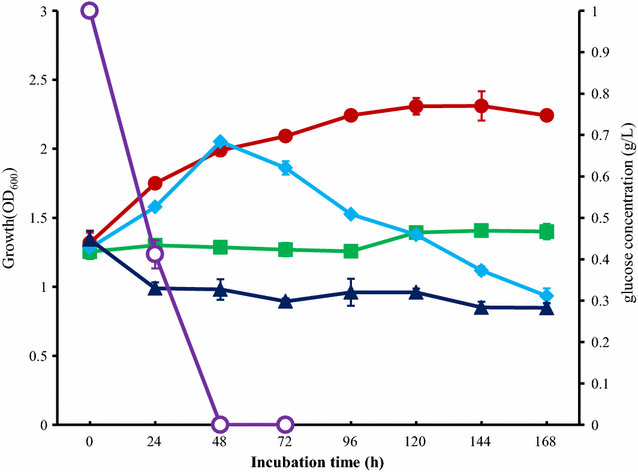
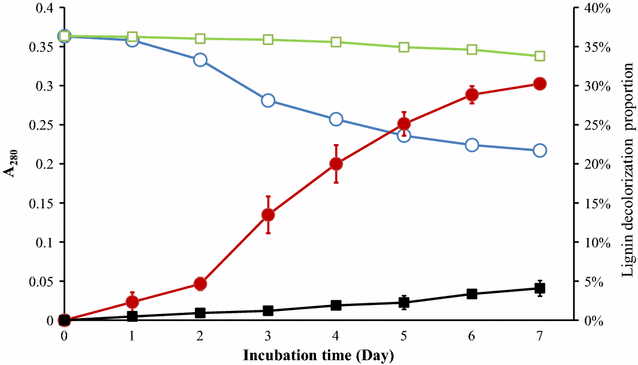
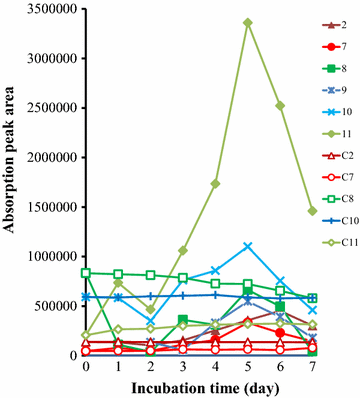
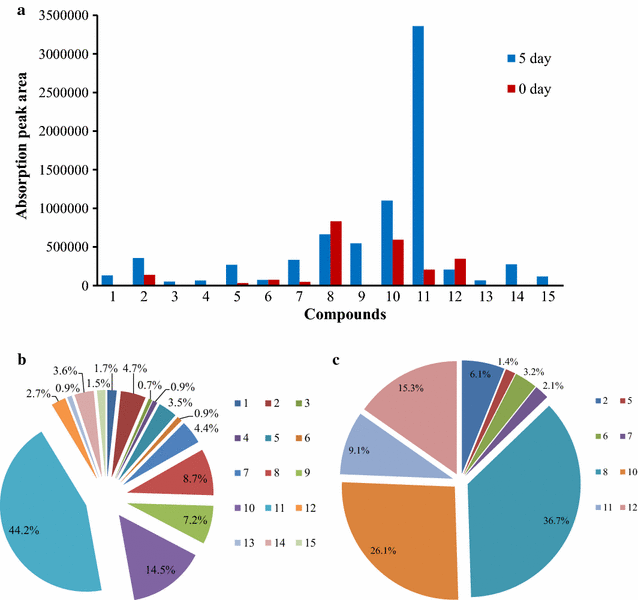
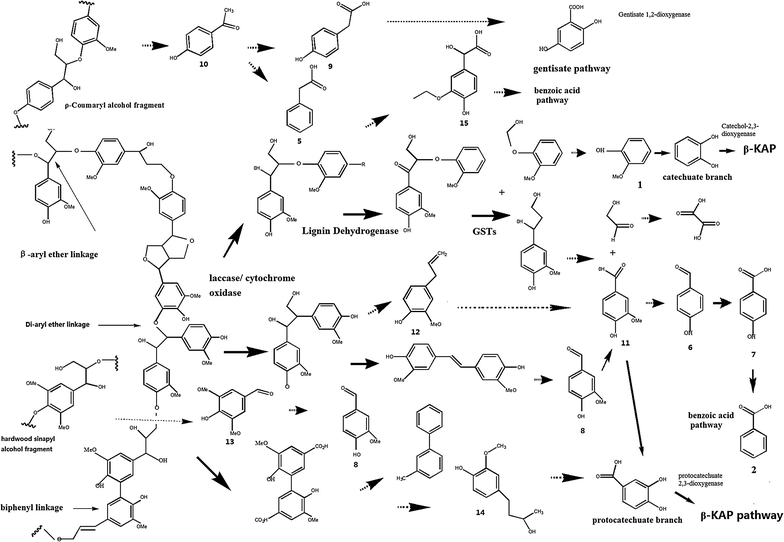
Results: To investigate the aromatic metabolites of strain L1 decomposing alkaline lignin, GC-MS analysis was performed and fifteen single phenol ring aromatic compounds were identified. The dominant absorption peak included phenylacetic acid, 4-hydroxy-benzoicacid, and vanillic acid with the highest proportion of metabolites resulting in 42%. Comparison proteomic analysis was carried out for further study showed that approximately 1447 kinds of proteins were produced, 141 of which were at least twofold up-regulated with alkaline lignin as the single carbon source. The up-regulated proteins contents different categories in the biological functions of protein including lignin degradation, ABC transport system, environmental response factors, protein synthesis, assembly, etc.
Conclusions: GC-MS analysis showed that alkaline lignin degradation of strain L1 produced 15 kinds of aromatic compounds. Comparison proteomic data and metabolic analysis showed that to ensure the degradation of lignin and growth of strain L1, multiple aspects of cells metabolism including transporter, environmental response factors, and protein synthesis were enhanced. Based on genome and proteomic analysis, at least four kinds of lignin degradation pathway might be present in strain L1, including a Gentisate pathway, the benzoic acid pathway and the β-ketoadipate pathway. The study provides an important basis for lignin degradation by bacteria.
Keywords: Alkaline lignin; Bacillus ligniniphilus L1; GC–MS; Proteomics.
Figures

Similar articles
-
Genomic analysis of Burkholderia sp. ISTR5 for biofunneling of lignin-derived compounds.Biotechnol Biofuels. 2019 Nov 27;12:277. doi: 10.1186/s13068-019-1606-5. eCollection 2019. Biotechnol Biofuels. 2019. PMID: 31788027 Free PMC article.
-
Characterization of Ligninolytic Bacteria and Analysis of Alkali-Lignin Biodegradation Products.Pol J Microbiol. 2020 Sep;69(3):339-347. doi: 10.33073/pjm-2020-037. Epub 2020 Sep 8. Pol J Microbiol. 2020. PMID: 33574863 Free PMC article.
-
Anaerobic Degradation of Syringic Acid by an Adapted Strain of Rhodopseudomonas palustris.Appl Environ Microbiol. 2020 Jan 21;86(3):e01888-19. doi: 10.1128/AEM.01888-19. Print 2020 Jan 21. Appl Environ Microbiol. 2020. PMID: 31732577 Free PMC article.
-
Aromatic metabolism of filamentous fungi in relation to the presence of aromatic compounds in plant biomass.Adv Appl Microbiol. 2015;91:63-137. doi: 10.1016/bs.aambs.2014.12.001. Epub 2015 Feb 24. Adv Appl Microbiol. 2015. PMID: 25911233 Review.
-
A comparison between the homocyclic aromatic metabolic pathways from plant-derived compounds by bacteria and fungi.Biotechnol Adv. 2019 Nov 15;37(7):107396. doi: 10.1016/j.biotechadv.2019.05.002. Epub 2019 May 7. Biotechnol Adv. 2019. PMID: 31075306 Review.
Cited by
-
Deconstruction of Lignin: From Enzymes to Microorganisms.Molecules. 2021 Apr 15;26(8):2299. doi: 10.3390/molecules26082299. Molecules. 2021. PMID: 33921125 Free PMC article. Review.
-
Recent Development of Extremophilic Bacteria and Their Application in Biorefinery.Front Bioeng Biotechnol. 2020 Jun 12;8:483. doi: 10.3389/fbioe.2020.00483. eCollection 2020. Front Bioeng Biotechnol. 2020. PMID: 32596215 Free PMC article. Review.
-
Production of Protocatechuic Acid from p-Hydroxyphenyl (H) Units and Related Aromatic Compounds Using an Aspergillus niger Cell Factory.mBio. 2021 Jun 29;12(3):e0039121. doi: 10.1128/mBio.00391-21. Epub 2021 Jun 22. mBio. 2021. PMID: 34154420 Free PMC article.
-
Lignin-Degrading Enzymes and the Potential of Pseudomonas putida as a Cell Factory for Lignin Degradation and Valorization.Microorganisms. 2025 Apr 18;13(4):935. doi: 10.3390/microorganisms13040935. Microorganisms. 2025. PMID: 40284771 Free PMC article. Review.
-
Isolation and Characterization of Lignocellulose-Degrading Geobacillus thermoleovorans from Yellowstone National Park.Appl Environ Microbiol. 2022 Jan 11;88(1):e0095821. doi: 10.1128/AEM.00958-21. Epub 2021 Oct 20. Appl Environ Microbiol. 2022. PMID: 34669438 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

