Nur77 suppresses hepatocellular carcinoma via switching glucose metabolism toward gluconeogenesis through attenuating phosphoenolpyruvate carboxykinase sumoylation
- PMID: 28240261
- PMCID: PMC5333363
- DOI: 10.1038/ncomms14420
Nur77 suppresses hepatocellular carcinoma via switching glucose metabolism toward gluconeogenesis through attenuating phosphoenolpyruvate carboxykinase sumoylation
Abstract
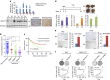
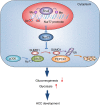
Gluconeogenesis, an essential metabolic process for hepatocytes, is downregulated in hepatocellular carcinoma (HCC). Here we show that the nuclear receptor Nur77 is a tumour suppressor for HCC that regulates gluconeogenesis. Low Nur77 expression in clinical HCC samples correlates with poor prognosis, and a Nur77 deficiency in mice promotes HCC development. Nur77 interacts with phosphoenolpyruvate carboxykinase (PEPCK1), the rate-limiting enzyme in gluconeogenesis, to increase gluconeogenesis and suppress glycolysis, resulting in ATP depletion and cell growth arrest. However, PEPCK1 becomes labile after sumoylation and is degraded via ubiquitination, which is augmented by the p300 acetylation of ubiquitin-conjugating enzyme 9 (Ubc9). Although Nur77 attenuates sumoylation and stabilizes PEPCK1 via impairing p300 activity and preventing the Ubc9-PEPCK1 interaction, Nur77 is silenced in HCC samples due to Snail-mediated DNA methylation of the Nur77 promoter. Our study reveals a unique mechanism to suppress HCC by switching from glycolysis to gluconeogenesis through Nur77 antagonism of PEPCK1 degradation.
Conflict of interest statement
The authors declare no competing financial interests.
Figures





References
-
- El-Serag H. B. & Rudolph K. L. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology 132, 2557–2576 (2007). - PubMed
-
- Forner A., Llovet J. M. & Bruix J. Hepatocellular carcinoma. Lancet 379, 1245–1255 (2012). - PubMed
-
- Marquardt J. U. & Thorgeirsson S. S. SnapShot: hepatocellular carcinoma. Cancer Cell 25, 550 e551 (2014). - PubMed
-
- Hanahan D. & Weinberg R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

