Genome-wide identification of splicing QTLs in the human brain and their enrichment among schizophrenia-associated loci
- PMID: 28240266
- PMCID: PMC5333373
- DOI: 10.1038/ncomms14519
Genome-wide identification of splicing QTLs in the human brain and their enrichment among schizophrenia-associated loci
Abstract
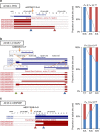
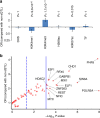
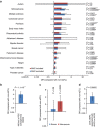
Detailed analyses of transcriptome have revealed complexity in regulation of alternative splicing (AS). These AS events often undergo modulation by genetic variants. Here we analyse RNA-sequencing data of prefrontal cortex from 206 individuals in combination with their genotypes and identify cis-acting splicing quantitative trait loci (sQTLs) throughout the genome. These sQTLs are enriched among exonic and H3K4me3-marked regions. Moreover, we observe significant enrichment of sQTLs among disease-associated loci identified by GWAS, especially in schizophrenia risk loci. Closer examination of each schizophrenia-associated loci revealed four regions (each encompasses NEK4, FXR1, SNAP91 or APOPT1), where the index SNP in GWAS is in strong linkage disequilibrium with sQTL SNP(s), suggesting dysregulation of AS as the underlying mechanism of the association signal. Our study provides an informative resource of sQTL SNPs in the human brain, which can facilitate understanding of the genetic architecture of complex brain disorders such as schizophrenia.
Conflict of interest statement
T.K. received a research grant from Takeda Pharmaceuticals Company Limited outside of this work. The remaining authors declare no competing financial interests.
Figures


References
-
- Barbosa-Morais N. L. et al. The evolutionary landscape of alternative splicing in vertebrate species. Science 338, 1587–1593 (2012). - PubMed
-
- Licatalosi D. D. & Darnell R. B. Splicing regulation in neurologic disease. Neuron 52, 93–101 (2006). - PubMed
-
- Raj B. & Blencowe B. J. Alternative splicing in the mammalian nervous system: recent insights into mechanisms and functional roles. Neuron 87, 14–27 (2015). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

