Role of DNA methylation in expression control of the IKZF3-GSDMA region in human epithelial cells
- PMID: 28241063
- PMCID: PMC5328393
- DOI: 10.1371/journal.pone.0172707
Role of DNA methylation in expression control of the IKZF3-GSDMA region in human epithelial cells
Abstract
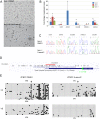
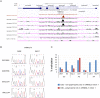
Chromosomal region 17q12-q21 is associated with asthma and harbors regulatory polymorphisms that influence expression levels of all five protein-coding genes in the region: IKAROS family zinc finger 3 (Aiolos) (IKZF3), zona pellucida binding protein 2 (ZPBP2), ORMDL sphingolipid biosynthesis regulator 3 (ORMDL3), and gasdermins A and B (GSDMA, GSDMB). Furthermore, DNA methylation in this region has been implicated as a potential modifier of the genetic risk of asthma development. To further characterize the effect of DNA methylation, we examined the impact of treatment with DNA methyltransferase inhibitor 5-aza-2'-deoxycytidine (5-aza-dC) that causes DNA demethylation, on expression and promoter methylation of the five 17q12-q21 genes in the human airway epithelium cell line NuLi-1, embryonic kidney epithelium cell line 293T and human adenocarcinoma cell line MCF-7. 5-aza-dC treatment led to upregulation of expression of GSDMA in all three cell lines. ZPBP2 was upregulated in NuLi-1, but remained repressed in 293T and MCF-7 cells, whereas ORMDL3 was upregulated in 293T and MCF-7 cells, but not NuLi-1. Upregulation of ZPBP2 and GSDMA was accompanied by a decrease in promoter methylation. Moreover, 5-aza-dC treatment modified allelic expression of ZPBP2 and ORMDL3 suggesting that different alleles may respond differently to treatment. We also identified a polymorphic CTCF-binding site in intron 1 of ORMDL3 carrying a CG SNP rs4065275 and determined its methylation level. The site's methylation was unaffected by 5-aza-dC treatment in NuLi-1 cells. We conclude that modest changes (8-13%) in promoter methylation levels of ZPBP2 and GSDMA may cause substantial changes in RNA levels and that allelic expression of ZPBP2 and ORMDL3 is mediated by DNA methylation.
Conflict of interest statement
Figures

References
-
- Schaefer S, Nadeau JH. The genetics of epigenetic inheritance: modes, molecules, and mechanisms. The Quarterly review of biology. 2015;90(4):381–415. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

