The transcriptome of HIV-1 infected intestinal CD4+ T cells exposed to enteric bacteria
- PMID: 28241075
- PMCID: PMC5344538
- DOI: 10.1371/journal.ppat.1006226
The transcriptome of HIV-1 infected intestinal CD4+ T cells exposed to enteric bacteria
Abstract
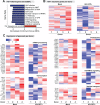
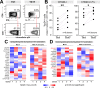
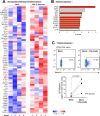
Global transcriptome studies can help pinpoint key cellular pathways exploited by viruses to replicate and cause pathogenesis. Previous data showed that laboratory-adapted HIV-1 triggers significant gene expression changes in CD4+ T cell lines and mitogen-activated CD4+ T cells from peripheral blood. However, HIV-1 primarily targets mucosal compartments during acute infection in vivo. Moreover, early HIV-1 infection causes extensive depletion of CD4+ T cells in the gastrointestinal tract that herald persistent inflammation due to the translocation of enteric microbes to the systemic circulation. Here, we profiled the transcriptome of primary intestinal CD4+ T cells infected ex vivo with transmitted/founder (TF) HIV-1. Infections were performed in the presence or absence of Prevotella stercorea, a gut microbe enriched in the mucosa of HIV-1-infected individuals that enhanced both TF HIV-1 replication and CD4+ T cell death ex vivo. In the absence of bacteria, HIV-1 triggered a cellular shutdown response involving the downregulation of HIV-1 reactome genes, while perturbing genes linked to OX40, PPAR and FOXO3 signaling. However, in the presence of bacteria, HIV-1 did not perturb these gene sets or pathways. Instead, HIV-1 enhanced granzyme expression and Th17 cell function, inhibited G1/S cell cycle checkpoint genes and triggered downstream cell death pathways in microbe-exposed gut CD4+ T cells. To gain insights on these differential effects, we profiled the gene expression landscape of HIV-1-uninfected gut CD4+ T cells exposed to bacteria. Microbial exposure upregulated genes involved in cellular proliferation, MAPK activation, Th17 cell differentiation and type I interferon signaling. Our findings reveal that microbial exposure influenced how HIV-1 altered the gut CD4+ T cell transcriptome, with potential consequences for HIV-1 susceptibility, cell survival and inflammation. The HIV-1- and microbe-altered pathways unraveled here may serve as a molecular blueprint to gain basic insights in mucosal HIV-1 pathogenesis.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
-
- Dalgleish AG, Beverley PC, Clapham PR, Crawford DH, Greaves MF, Weiss RA. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312(5996):763–7. - PubMed
-
- Jekle A, Keppler OT, De Clercq E, Schols D, Weinstein M, Goldsmith MA. In vivo evolution of human immunodeficiency virus type 1 toward increased pathogenicity through CXCR4-mediated killing of uninfected CD4 T cells. Journal of virology. 2003;77(10):5846–54. 10.1128/JVI.77.10.5846-5854.2003 - DOI - PMC - PubMed
-
- Ochsenbauer C, Edmonds TG, Ding H, Keele BF, Decker J, Salazar MG, et al. Generation of transmitted/founder HIV-1 infectious molecular clones and characterization of their replication capacity in CD4 T lymphocytes and monocyte-derived macrophages. Journal of virology. 2012;86(5):2715–28. Epub 2011/12/23. 10.1128/JVI.06157-11 - DOI - PMC - PubMed
-
- Salazar-Gonzalez JF, Salazar MG, Keele BF, Learn GH, Giorgi EE, Li H, et al. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. The Journal of experimental medicine. 2009;206(6):1273–89. Epub 2009/06/03. 10.1084/jem.20090378 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

