Exome Sequencing in a Family with Luminal-Type Breast Cancer Underpinned by Variation in the Methylation Pathway
- PMID: 28241424
- PMCID: PMC5343999
- DOI: 10.3390/ijms18020467
Exome Sequencing in a Family with Luminal-Type Breast Cancer Underpinned by Variation in the Methylation Pathway
Abstract
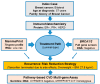
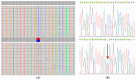
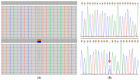
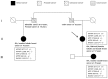
Panel-based next generation sequencing (NGS) is currently preferred over whole exome sequencing (WES) for diagnosis of familial breast cancer, due to interpretation challenges caused by variants of uncertain clinical significance (VUS). There is also no consensus on the selection criteria for WES. In this study, a pathology-supported genetic testing (PSGT) approach was used to select two BRCA1/2 mutation-negative breast cancer patients from the same family for WES. Homozygosity for the MTHFR 677 C>T mutation detected during this PSGT pre-screen step was considered insufficient to cause bilateral breast cancer in the index case and her daughter diagnosed with early-onset breast cancer (<30 years). Extended genetic testing using WES identified the RAD50 R385C missense mutation in both cases. This rare variant with a minor allele frequency (MAF) of <0.001 was classified as a VUS after exclusion in an affected cousin and extended genotyping in 164 unrelated breast cancer patients and 160 controls. Detection of functional polymorphisms (MAF > 5%) in the folate pathway in all three affected family members is consistent with inheritance of the luminal-type breast cancer in the family. PSGT assisted with the decision to pursue extended genetic testing and facilitated clinical interpretation of WES aimed at reduction of recurrence risk.
Keywords: breast cancer; exome sequencing; methylation pathway; pathology; pharmacogenomics.
Conflict of interest statement
Maritha J. Kotze is a director and shareholder of Gknowmix (Pty) Ltd. that has developed a database tool for research translation under the auspices of the South African Medical Research Council. Susan J. van Rensburg is a scientific advisor of Gknowmix, with no potential conflict of interest reported by the other authors. Maritha J. Kotze is the inventor of a South African Medical Research Council patent 2001/5419 (2 July 2001) entitled “A method of diagnosing patients with cardiovascular disease or a genetic predisposition for cardiovascular disease”. The funding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
Figures

Similar articles
-
BRCA1 and BRCA2 Testing through Next Generation Sequencing in a Small Cohort of Italian Breast/Ovarian Cancer Patients: Novel Pathogenic and Unknown Clinical Significance Variants.Int J Mol Sci. 2019 Jul 12;20(14):3442. doi: 10.3390/ijms20143442. Int J Mol Sci. 2019. PMID: 31336956 Free PMC article.
-
Whole-exome sequencing and targeted gene sequencing provide insights into the role of PALB2 as a male breast cancer susceptibility gene.Cancer. 2017 Jan 1;123(2):210-218. doi: 10.1002/cncr.30337. Epub 2016 Sep 20. Cancer. 2017. PMID: 27648926
-
Genomic medicine and risk prediction across the disease spectrum.Crit Rev Clin Lab Sci. 2015;52(3):120-37. doi: 10.3109/10408363.2014.997930. Epub 2015 Jan 19. Crit Rev Clin Lab Sci. 2015. PMID: 25597499 Review.
-
Frequency of pathogenic germline mutation in CHEK2, PALB2, MRE11, and RAD50 in patients at high risk for hereditary breast cancer.Breast Cancer Res Treat. 2017 Jan;161(1):95-102. doi: 10.1007/s10549-016-4034-2. Epub 2016 Oct 25. Breast Cancer Res Treat. 2017. PMID: 27783279
-
Prevalence of pathogenic variants and variants of unknown significance in patients at high risk of breast cancer: A systematic review and meta-analysis of gene-panel data.Crit Rev Oncol Hematol. 2018 Dec;132:138-144. doi: 10.1016/j.critrevonc.2018.09.009. Epub 2018 Sep 14. Crit Rev Oncol Hematol. 2018. PMID: 30447919
Cited by
-
Genetics of breast cancer in African populations: a literature review.Glob Health Epidemiol Genom. 2018 May 11;3:e8. doi: 10.1017/gheg.2018.8. eCollection 2018. Glob Health Epidemiol Genom. 2018. PMID: 30263132 Free PMC article. Review.
-
Identification of an iron-responsive subtype in two children diagnosed with relapsing-remitting multiple sclerosis using whole exome sequencing.Mol Genet Metab Rep. 2019 Mar 23;19:100465. doi: 10.1016/j.ymgmr.2019.100465. eCollection 2019 Jun. Mol Genet Metab Rep. 2019. PMID: 30963028 Free PMC article.
-
Human whole genome sequencing in South Africa.Sci Rep. 2021 Jan 12;11(1):606. doi: 10.1038/s41598-020-79794-x. Sci Rep. 2021. PMID: 33436733 Free PMC article.
-
Whole Exome Sequencing in South Africa: Stakeholder Views on Return of Individual Research Results and Incidental Findings.Front Genet. 2022 Jun 8;13:864822. doi: 10.3389/fgene.2022.864822. eCollection 2022. Front Genet. 2022. PMID: 35754817 Free PMC article.
-
Implementation of multigene panel testing for breast and ovarian cancer in South Africa: A step towards excellence in oncology for the public sector.Front Oncol. 2022 Dec 7;12:938561. doi: 10.3389/fonc.2022.938561. eCollection 2022. Front Oncol. 2022. PMID: 36568162 Free PMC article.
References
-
- Nordgard S.H., Alnaes G.I., Hihn B., Lingjaerde O.C., Liestøl K., Tsalenko A., Sørlie T., Lønning P.E., Børresen-Dale A.L., Kristensen V.N. Pathway based analysis of SNPs with relevance to 5-FU therapy: Relation to intratumoral mRNA expression and survival. Int. J. Cancer. 2008;123:577–585. doi: 10.1002/ijc.23541. - DOI - PubMed
-
- Babyshkina N., Malinovskaya E., Nazarenko M., Koval M., Gervas P., Potapova O., Slonimskaya E., Cherdyntseva N. The effect of folate-related SNPs on clinicopathological features, response to neoadjuvant treatment and survival in pre- and postmenopausal breast cancer patients. Gene. 2013;518:397–404. doi: 10.1016/j.gene.2012.12.095. - DOI - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

