Mapping Post-Transcriptional Modifications onto Transfer Ribonucleic Acid Sequences by Liquid Chromatography Tandem Mass Spectrometry
- PMID: 28241457
- PMCID: PMC5372733
- DOI: 10.3390/biom7010021
Mapping Post-Transcriptional Modifications onto Transfer Ribonucleic Acid Sequences by Liquid Chromatography Tandem Mass Spectrometry
Abstract
Liquid chromatography, coupled with tandem mass spectrometry, has become one of the most popular methods for the analysis of post-transcriptionally modified transfer ribonucleic acids (tRNAs). Given that the information collected using this platform is entirely determined by the mass of the analyte, it has proven to be the gold standard for accurately assigning nucleobases to the sequence. For the past few decades many labs have worked to improve the analysis, contiguous to instrumentation manufacturers developing faster and more sensitive instruments. With biological discoveries relating to ribonucleic acid happening more frequently, mass spectrometry has been invaluable in helping to understand what is happening at the molecular level. Here we present a brief overview of the methods that have been developed and refined for the analysis of modified tRNAs by liquid chromatography tandem mass spectrometry.
Keywords: modified nucleosides; LC‐MS/MS; RNA sequencing; tRNA; tandem mass spectrometry.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
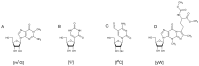
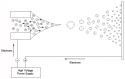




Similar articles
-
Sequence mapping of transfer RNA chemical modifications by liquid chromatography tandem mass spectrometry.Methods. 2016 Sep 1;107:73-8. doi: 10.1016/j.ymeth.2016.03.016. Epub 2016 Mar 24. Methods. 2016. PMID: 27033178 Free PMC article.
-
Metabolic de-isotoping for improved LC-MS characterization of modified RNAs.J Am Soc Mass Spectrom. 2014 Jul;25(7):1114-23. doi: 10.1007/s13361-014-0889-9. Epub 2014 Apr 24. J Am Soc Mass Spectrom. 2014. PMID: 24760295
-
Identification and Quantification of (t)RNA Modifications in Pseudomonas aeruginosa by Liquid Chromatography-Tandem Mass Spectrometry.Chembiochem. 2019 Jun 3;20(11):1430-1437. doi: 10.1002/cbic.201800741. Epub 2019 Apr 5. Chembiochem. 2019. PMID: 30644616
-
[Current advances in the analysis of free RNA modified nucleosides by high performance liquid chromatography-tandem mass spectrometry].Se Pu. 2025 Jan;43(1):3-12. doi: 10.3724/SP.J.1123.2024.07004. Se Pu. 2025. PMID: 39722616 Free PMC article. Review. Chinese.
-
Characterizing RNA modifications in the central nervous system and single cells by RNA sequencing and liquid chromatography-tandem mass spectrometry techniques.Anal Bioanal Chem. 2023 Jul;415(18):3739-3748. doi: 10.1007/s00216-023-04604-y. Epub 2023 Feb 25. Anal Bioanal Chem. 2023. PMID: 36840809 Review.
Cited by
-
Comprehensive nucleoside analysis of archaeal RNA modification profiles reveals an m7G in the conserved P loop of 23S rRNA.Cell Rep. 2025 Apr 22;44(4):115471. doi: 10.1016/j.celrep.2025.115471. Epub 2025 Mar 24. Cell Rep. 2025. PMID: 40131932 Free PMC article.
-
Stress-induced modification of Escherichia coli tRNA generates 5-methylcytidine in the variable loop.Proc Natl Acad Sci U S A. 2024 Nov 12;121(46):e2317857121. doi: 10.1073/pnas.2317857121. Epub 2024 Nov 4. Proc Natl Acad Sci U S A. 2024. PMID: 39495928 Free PMC article.
-
Improved RNA modification mapping of cellular non-coding RNAs using C- and U-specific RNases.Analyst. 2020 Feb 3;145(3):816-827. doi: 10.1039/c9an02111f. Analyst. 2020. PMID: 31825413 Free PMC article.
-
Radical Transfer Dissociation for De Novo Characterization of Modified Ribonucleic Acids by Mass Spectrometry.Angew Chem Int Ed Engl. 2020 Mar 9;59(11):4309-4313. doi: 10.1002/anie.201914275. Epub 2020 Jan 31. Angew Chem Int Ed Engl. 2020. PMID: 31867820 Free PMC article.
-
Ionizing radiation and chemical oxidant exposure impacts on Cryptococcus neoformans transfer RNAs.PLoS One. 2022 Mar 29;17(3):e0266239. doi: 10.1371/journal.pone.0266239. eCollection 2022. PLoS One. 2022. PMID: 35349591 Free PMC article.
References
-
- Machnicka M.A., Milanowska K., Osman Oglou O., Purta E., Kurkowska M., Olchowik A., Januszewski W., Kalinowski S., Dunin-Horkawicz S., Rother K.M., et al. Modomics: A database of RNA modification pathways—2013 update. Nucleic Acids Res. 2013;41:D262–D267. doi: 10.1093/nar/gks1007. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

