Accurate and equitable medical genomic analysis requires an understanding of demography and its influence on sample size and ratio
- PMID: 28241850
- PMCID: PMC5330117
- DOI: 10.1186/s13059-017-1172-8
Accurate and equitable medical genomic analysis requires an understanding of demography and its influence on sample size and ratio
Abstract
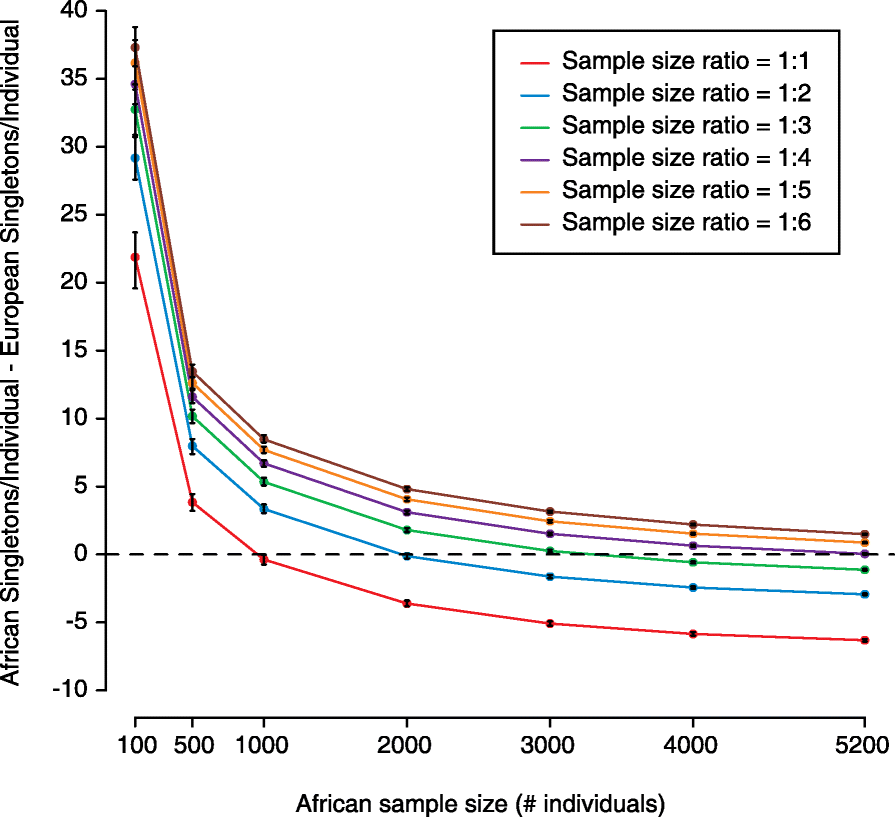
In a recent study, Petrovski and Goldstein reported that (non-Finnish) Europeans have significantly fewer nonsynonymous singletons in Online Mendelian Inheritance in Man (OMIM) disease genes compared with Africans, Latinos, South Asians, East Asians, and other unassigned non-Europeans. We use simulations of Exome Aggregation Consortium (ExAC) data to show that sample size and ratio interact to influence the number of these singletons identified in a cohort. These interactions are different across ancestries and can lead to the same number of identified singletons in both Europeans and non-Europeans without an equal number of samples. We conclude that there is a need to account for the ancestry-specific influence of demography on genomic architecture and rare variant analysis in order to address inequalities in medical genomic analysis.The authors of the original article were invited to submit a response, but declined to do so. Please see related Open Letter: http://genomebiology.biomedcentral.com/articles/10.1186/s13059-016-1016-y.
Figures

Comment on
-
Unequal representation of genetic variation across ancestry groups creates healthcare inequality in the application of precision medicine.Genome Biol. 2016 Jul 14;17(1):157. doi: 10.1186/s13059-016-1016-y. Genome Biol. 2016. PMID: 27418169 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

