Divergent Effects of miR-181 Family Members on Myocardial Function Through Protective Cytosolic and Detrimental Mitochondrial microRNA Targets
- PMID: 28242633
- PMCID: PMC5524005
- DOI: 10.1161/JAHA.116.004694
Divergent Effects of miR-181 Family Members on Myocardial Function Through Protective Cytosolic and Detrimental Mitochondrial microRNA Targets
Abstract
Background: MicroRNA (miRNA) is a type of noncoding RNA that can repress the expression of target genes through posttranscriptional regulation. In addition to numerous physiologic roles for miRNAs, they play an important role in pathophysiologic processes affecting cardiovascular health. Previously, we reported that nuclear encoded microRNA (miR-181c) is present in heart mitochondria, and importantly, its overexpression affects mitochondrial function by regulating mitochondrial gene expression.
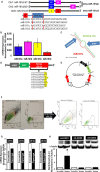
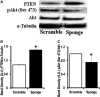
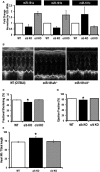
Methods and results: To investigate further how the miR-181 family affects the heart, we suppressed miR-181 using a miR-181-sponge containing 10 repeated complementary miR-181 "seed" sequences and generated a set of H9c2 cells, a cell line derived from rat myoblast, by stably expressing either a scrambled or miR-181-sponge sequence. Sponge-H9c2 cells showed a decrease in reactive oxygen species production and reduced basal mitochondrial respiration and protection against doxorubicin-induced oxidative stress. We also found that miR-181a/b targets phosphatase and tensin homolog (PTEN), and the sponge-expressing stable cells had increased PTEN activity and decreased PI3K signaling. In addition, we have used miR-181a/b-/- and miR-181c/d-/- knockout mice and subjected them to ischemia-reperfusion injury. Our results suggest divergent effects of different miR-181 family members: miR-181a/b targets PTEN in the cytosol, resulting in an increase in infarct size in miR-181a/b-/- mice due to increased PTEN signaling, whereas miR-181c targets mt-COX1 in the mitochondria, resulting in decreased infarct size in miR-181c/d-/- mice.
Conclusions: The miR-181 family alters the myocardial response to oxidative stress, notably with detrimental effects by targeting mt-COX1 (miR-181c) or with protection by targeting PTEN (miR-181a/b).
Keywords: PI3 kinase; miR‐181; microRNA; mitochondria; mitochondrial miRNA; mitochondrial respiratory complex IV; mt‐COX1; oxidative stress; phosphatase and tensin homolog; reperfusion injury.
© 2017 The Authors. Published on behalf of the American Heart Association, Inc., by Wiley Blackwell.
Figures






References
-
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

